| Sequence ID | dm3.chr3L |
|---|---|
| Location | 5,225,384 – 5,225,477 |
| Length | 93 |
| Max. P | 0.640572 |

| Location | 5,225,384 – 5,225,477 |
|---|---|
| Length | 93 |
| Sequences | 11 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 77.22 |
| Shannon entropy | 0.44355 |
| G+C content | 0.41036 |
| Mean single sequence MFE | -17.52 |
| Consensus MFE | -11.31 |
| Energy contribution | -11.41 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.640572 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
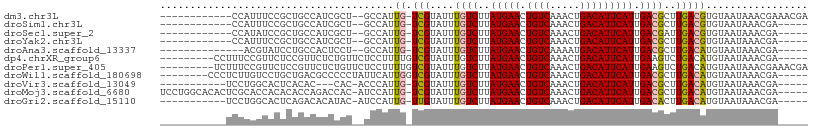
>dm3.chr3L 5225384 93 + 24543557 ------------CCAUUUCCGCUGCCAUCGCU--GCCAUUG-UCGUAUUUGUCUUAUGAACUGUCAAACUGACAUUCAUUGACGCUUGACGUGUAAUAAACGAAACGA ------------...((((.((.((....)).--))(((.(-(((....((((..(((((.((((.....))))))))).))))..)))))))........))))... ( -18.80, z-score = -0.88, R) >droSim1.chr3L 4744134 88 + 22553184 ------------CCAUUUCCGCUGCCAUCGCU--GCCAUUG-UCGUAUUUGUCUUAUGAACUGUCAAACUGACAUUCAUUGACGCUUGACGUGUAAUAAACGA----- ------------........((.((....)).--))(((.(-(((....((((..(((((.((((.....))))))))).))))..)))))))..........----- ( -17.80, z-score = -1.01, R) >droSec1.super_2 5178744 88 + 7591821 ------------CCAUAUCCGCUGCCAUCGCU--GCCAUUG-UCGUAUUUGUCUUAUGAACUGUCAAACUGACAUUCAUUGACGAUUGACGUGUAAUAAACGA----- ------------........((.((....)).--))(((.(-(((...(((((..(((((.((((.....))))))))).))))).)))))))..........----- ( -20.10, z-score = -1.67, R) >droYak2.chr3L 5788546 88 + 24197627 ------------CCAUUUCCGCUGCCAUCGCU--GCCAUUG-UCGUAUUUGUCUUAUGAACUGUCAAACUGACAUUCAUUGACGCUUGACGUGUAAUAAACGA----- ------------........((.((....)).--))(((.(-(((....((((..(((((.((((.....))))))))).))))..)))))))..........----- ( -17.80, z-score = -1.01, R) >droAna3.scaffold_13337 9982757 86 - 23293914 --------------ACGUAUCCUGCCACUCCU--GCCAUUG-UCGUAUUUGUCUUAUGAACUGUCAAAAUGACAUUCAUUGACGCUUGACAUGUAAUAAACGA----- --------------.(((.....((.......--))...((-(((....((((..(((((.((((.....))))))))).))))..)))))........))).----- ( -16.60, z-score = -1.53, R) >dp4.chrXR_group6 7481980 94 + 13314419 ---------CCUUUCCGUUCUCCGUUCUCUGUUCUCCUUUUGUCGUAUUUGUCUUAUGAACUGUCAAACUGACAUUCAUUGAAGUCUGACAUGUAAUAAACGA----- ---------....................((((.......(((((.((...((..(((((.((((.....))))))))).)).)).))))).......)))).----- ( -13.24, z-score = -0.60, R) >droPer1.super_405 8644 99 + 24988 ---------UCUUUCCGUUCUCCGUUCUCUGUUCUCCUUUUGUCGUAUUUGUCUUAUGAACUGUCAAACUGACAUUCAUUGAAGUCUGACAUGUAAUAAACGAAACGA ---------.............((((.((.(((.......(((((.((...((..(((((.((((.....))))))))).)).)).))))).......))))))))). ( -16.44, z-score = -0.96, R) >droWil1.scaffold_180698 6935464 95 - 11422946 --------CCCUCUUGUCCUGCUGACGCCCCCUAUUCAUUGGUCGUAUUUGUCUUAUGAACUGUCAAACUGACAUUCAUUGACGCUUGACAUGUAAUAAACGA----- --------......((((..((....((..((........))..))....(((..(((((.((((.....))))))))).)))))..))))............----- ( -20.20, z-score = -2.63, R) >droVir3.scaffold_13049 7963826 87 + 25233164 -----------UCCUGGCACUCACAC---CAC-ACCCAUUG-UCGUAUUUGUCUUAUGAACUGUCAAACUGACAUUCAUUGACGCUUGACAUGUAAUAAACGA----- -----------...(((........)---)).-......((-(((....((((..(((((.((((.....))))))))).))))..)))))............----- ( -16.40, z-score = -1.04, R) >droMoj3.scaffold_6680 11626371 101 + 24764193 UCCUGGCACACUCGCACCACACACCAGACCAC-AUCCAUUG-UCGUAUUUGUCUUAUGAACUGUCAAACUGACAUUCAUUGACGCUUGACAUGUAAUAAACGA----- ..((((.................)))).....-......((-(((....((((..(((((.((((.....))))))))).))))..)))))............----- ( -18.33, z-score = -0.55, R) >droGri2.scaffold_15110 7016550 90 + 24565398 -----------UCCUGGCACUCAGACACAUAC-AUCCAUUG-UUGUAUUUGUCUUAUGAACUGUCAAACUGACAUUCAUUGACACUUGACAUGUAAUAAACGA----- -----------.....(((.(((((((.((((-(.......-.))))).))))..(((((.((((.....))))))))).......)))..))).........----- ( -17.00, z-score = -0.58, R) >consensus ___________UCCAUGUCCGCUGCCAUCCCU__GCCAUUG_UCGUAUUUGUCUUAUGAACUGUCAAACUGACAUUCAUUGACGCUUGACAUGUAAUAAACGA_____ ............................................((...((((..(((((.((((.....))))))))).))))....)).................. (-11.31 = -11.41 + 0.10)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:04:34 2011