
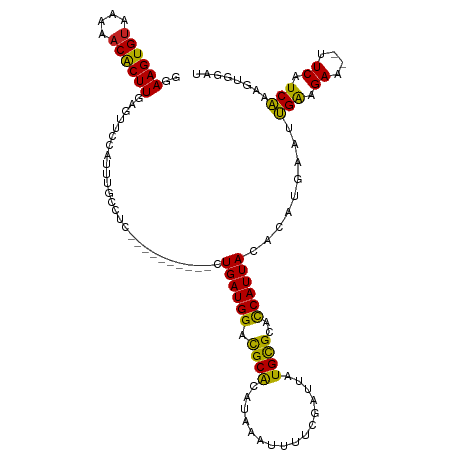
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 5,211,692 – 5,211,797 |
| Length | 105 |
| Max. P | 0.899297 |

| Location | 5,211,692 – 5,211,797 |
|---|---|
| Length | 105 |
| Sequences | 11 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 79.17 |
| Shannon entropy | 0.40716 |
| G+C content | 0.38006 |
| Mean single sequence MFE | -26.26 |
| Consensus MFE | -13.74 |
| Energy contribution | -12.94 |
| Covariance contribution | -0.81 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.777072 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 5211692 105 + 24543557 GGAAGUGUAAAAACACUUGAGUUCCAUUUGCCUC----------CUGAUGGACGCACAUAAAUUUUCGAUUAUGCGCACCAUUAGACAUGAAUUGAAGAA--UUCAUCAAAGUGGAU ..((((((....))))))....(((((((.....----------(((((((..((.(((((........))))).)).)))))))..(((((((....))--)))))..))))))). ( -29.40, z-score = -3.16, R) >droSim1.chr3L 4735090 105 + 22553184 GGAAGUGUAAAAACACUUGAGUUCCAUUUGCCUC----------CUGAUGGACGCACAUAAAUUUUCGAUUAUGCGCACCAUUAGACAUGAAUUGAAGAA--UUCAUCAAAGUGGAU ..((((((....))))))....(((((((.....----------(((((((..((.(((((........))))).)).)))))))..(((((((....))--)))))..))))))). ( -29.40, z-score = -3.16, R) >droSec1.super_2 5165104 105 + 7591821 GGAAGUGCAAAAACACUUGAGUUCCAUUUGCCUC----------CUGAUGGACGCAAAUAAAUUUUCGAUUAUGUGCACCAUUAGACAUGAAUUGAAGAA--UUCAUCAAAUUGGAU ..(((((......)))))((((((.((((((.((----------(....))).)))))).....(((((((((((..........))))).)))))))))--)))............ ( -25.00, z-score = -1.80, R) >droYak2.chr3L 5774561 105 + 24197627 GGAAGUGUAAAAACACUUGAGUUCCAUUUGCCUC----------CUGAUGGACGCACAUAAAUUUUCGAUUAUGCGCACCAUUAGACAUGAAUUGAAGAA--UUCAUCAAAGUGGAU ..((((((....))))))....(((((((.....----------(((((((..((.(((((........))))).)).)))))))..(((((((....))--)))))..))))))). ( -29.40, z-score = -3.16, R) >droEre2.scaffold_4784 7906445 105 + 25762168 GGAAGUGUAAAAACACUUGAGUUCCAUUUGCCUC----------CUGAUGGACGCACAUAAAUUUUCGAUUAUGCGCACCAUUAGACAUGAAUUGAAGAA--UUCAUCAAAGUGGAU ..((((((....))))))....(((((((.....----------(((((((..((.(((((........))))).)).)))))))..(((((((....))--)))))..))))))). ( -29.40, z-score = -3.16, R) >droAna3.scaffold_13337 9967490 105 - 23293914 GGAAGUGUAAAGACACUUGAAUUCCAUUUGCCUU----------CUGAUGGACGCGCAUAAAUUUUCGAUUAUGCGCACCAUUACACACGAAUUGAGGAA--UUCAUCAAAGUGGAU ..((((((....))))))((((((((((((....----------.((((((..((((((((........)))))))).))))))....)))))...))))--)))............ ( -31.10, z-score = -3.42, R) >dp4.chrXR_group6 7462425 111 + 13314419 GGAAGUGUAAAAACACUUGAAUUCCAUUUUGCUUUGCUUUUGUGCUGAUGGACGCACAUAAAUUUUCGAUUAUGCGGACCAUUACACAGGAAUUGAAGAAAAUUCAUCAGA------ ..((((((....)))))).((((((.....((...))...((((.((((((.((((..(((........)))))))..))))))))))))))))(((.....)))......------ ( -25.00, z-score = -1.47, R) >droPer1.super_2387 260 111 - 5444 GGAAGUGUAAAAACACUUGAAUUCCAUUUUGCUUUGCUUUUGUGCUGAUGGACGCACAUAAAUUUUCGAUUAUGCGGACCAUUACACAGGAAUUGAAGAAAAUUCAUCAGA------ ..((((((....)))))).((((((.....((...))...((((.((((((.((((..(((........)))))))..))))))))))))))))(((.....)))......------ ( -25.00, z-score = -1.47, R) >droVir3.scaffold_13049 7945016 101 + 25233164 UAAAGUGUAAA-ACGCUUGAAAACCAUUUGGCUUU---------UUGAUGGGCGCACAUAAAUUUGCGAUUAUGCGGUUCAUUACAUAA----CGAAGAGCUCUCAUCGAUAUAA-- ..(((((....-.)))))(((((((....)).)))---------))((((((.((.((......))((.(((((..(....)..)))))----))....)).)))))).......-- ( -19.40, z-score = -0.10, R) >droMoj3.scaffold_6680 11602535 102 + 24764193 UGAAGUGUAAA-ACGCUUGAGAACCAUUUGGCUGU---------UUGAUGGGCGCACAUAAAUUUGCGAUUAUGCGGUUCAUUACAUAA----CGAAGAGCACUCAUCGAUGUAAA- (.(((((....-.))))).)(((((.....(((..---------......)))(((........)))........))))).((((((..----(((.((....)).))))))))).- ( -22.60, z-score = -0.06, R) >droGri2.scaffold_15110 6999775 101 + 24565398 UGAAGUGUAAA-ACGCUUGAAAGCCAUUUGACUUU---------UUGAUGGGUGCACAUAAAUUUACGAUUAUGCGGCCCAUUACAUAA----CGGAGAGCUCUCAUCGCUGUAA-- ..(((((....-.)))))((.(((..((((.....---------.((((((((.(.(((((........))))).))))))))).....----))))..))).))..........-- ( -23.20, z-score = -0.72, R) >consensus GGAAGUGUAAAAACACUUGAGUUCCAUUUGCCUC__________CUGAUGGACGCACAUAAAUUUUCGAUUAUGCGCACCAUUACACAUGAAUUGAAGAA__UUCAUCAAAGUGGAU ..((((((....))))))...........................((((((.((((................))))..))))))........((((.((....)).))))....... (-13.74 = -12.94 + -0.81)

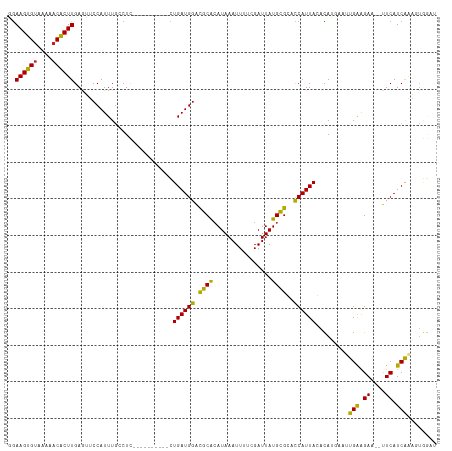
| Location | 5,211,692 – 5,211,797 |
|---|---|
| Length | 105 |
| Sequences | 11 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 79.17 |
| Shannon entropy | 0.40716 |
| G+C content | 0.38006 |
| Mean single sequence MFE | -27.25 |
| Consensus MFE | -9.65 |
| Energy contribution | -9.45 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.741734 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 5211692 105 - 24543557 AUCCACUUUGAUGAA--UUCUUCAAUUCAUGUCUAAUGGUGCGCAUAAUCGAAAAUUUAUGUGCGUCCAUCAG----------GAGGCAAAUGGAACUCAAGUGUUUUUACACUUCC .((((.(((((((((--((....))))))).(((.(((((((((((((........))))))))).)))).))----------)...))))))))....((((((....)))))).. ( -31.70, z-score = -3.90, R) >droSim1.chr3L 4735090 105 - 22553184 AUCCACUUUGAUGAA--UUCUUCAAUUCAUGUCUAAUGGUGCGCAUAAUCGAAAAUUUAUGUGCGUCCAUCAG----------GAGGCAAAUGGAACUCAAGUGUUUUUACACUUCC .((((.(((((((((--((....))))))).(((.(((((((((((((........))))))))).)))).))----------)...))))))))....((((((....)))))).. ( -31.70, z-score = -3.90, R) >droSec1.super_2 5165104 105 - 7591821 AUCCAAUUUGAUGAA--UUCUUCAAUUCAUGUCUAAUGGUGCACAUAAUCGAAAAUUUAUUUGCGUCCAUCAG----------GAGGCAAAUGGAACUCAAGUGUUUUUGCACUUCC .((((.(((((((((--((....))))))).(((.((((((((.((((........)))).)))).)))).))----------)...))))))))....((((((....)))))).. ( -26.40, z-score = -2.22, R) >droYak2.chr3L 5774561 105 - 24197627 AUCCACUUUGAUGAA--UUCUUCAAUUCAUGUCUAAUGGUGCGCAUAAUCGAAAAUUUAUGUGCGUCCAUCAG----------GAGGCAAAUGGAACUCAAGUGUUUUUACACUUCC .((((.(((((((((--((....))))))).(((.(((((((((((((........))))))))).)))).))----------)...))))))))....((((((....)))))).. ( -31.70, z-score = -3.90, R) >droEre2.scaffold_4784 7906445 105 - 25762168 AUCCACUUUGAUGAA--UUCUUCAAUUCAUGUCUAAUGGUGCGCAUAAUCGAAAAUUUAUGUGCGUCCAUCAG----------GAGGCAAAUGGAACUCAAGUGUUUUUACACUUCC .((((.(((((((((--((....))))))).(((.(((((((((((((........))))))))).)))).))----------)...))))))))....((((((....)))))).. ( -31.70, z-score = -3.90, R) >droAna3.scaffold_13337 9967490 105 + 23293914 AUCCACUUUGAUGAA--UUCCUCAAUUCGUGUGUAAUGGUGCGCAUAAUCGAAAAUUUAUGCGCGUCCAUCAG----------AAGGCAAAUGGAAUUCAAGUGUCUUUACACUUCC ...........((((--((((........(.((..(((((((((((((........))))))))).)))))).----------)........))))))))(((((....)))))... ( -30.59, z-score = -3.35, R) >dp4.chrXR_group6 7462425 111 - 13314419 ------UCUGAUGAAUUUUCUUCAAUUCCUGUGUAAUGGUCCGCAUAAUCGAAAAUUUAUGUGCGUCCAUCAGCACAAAAGCAAAGCAAAAUGGAAUUCAAGUGUUUUUACACUUCC ------...((.((....)).))(((((((((((.((((.((((((((........))))))).).))))..)))))...((...)).....)))))).((((((....)))))).. ( -26.60, z-score = -2.86, R) >droPer1.super_2387 260 111 + 5444 ------UCUGAUGAAUUUUCUUCAAUUCCUGUGUAAUGGUCCGCAUAAUCGAAAAUUUAUGUGCGUCCAUCAGCACAAAAGCAAAGCAAAAUGGAAUUCAAGUGUUUUUACACUUCC ------...((.((....)).))(((((((((((.((((.((((((((........))))))).).))))..)))))...((...)).....)))))).((((((....)))))).. ( -26.60, z-score = -2.86, R) >droVir3.scaffold_13049 7945016 101 - 25233164 --UUAUAUCGAUGAGAGCUCUUCG----UUAUGUAAUGAACCGCAUAAUCGCAAAUUUAUGUGCGCCCAUCAA---------AAAGCCAAAUGGUUUUCAAGCGU-UUUACACUUUA --.......((((.(.((..((((----((....)))))).(((((((........))))))).)))))))..---------((((((....)))))).......-........... ( -18.70, z-score = -0.54, R) >droMoj3.scaffold_6680 11602535 102 - 24764193 -UUUACAUCGAUGAGUGCUCUUCG----UUAUGUAAUGAACCGCAUAAUCGCAAAUUUAUGUGCGCCCAUCAA---------ACAGCCAAAUGGUUCUCAAGCGU-UUUACACUUCA -........((((.((((..((((----((....))))))..((((((........)))))))))).))))..---------..((((....)))).........-........... ( -19.60, z-score = -0.62, R) >droGri2.scaffold_15110 6999775 101 - 24565398 --UUACAGCGAUGAGAGCUCUCCG----UUAUGUAAUGGGCCGCAUAAUCGUAAAUUUAUGUGCACCCAUCAA---------AAAGUCAAAUGGCUUUCAAGCGU-UUUACACUUCA --.....((..((((((((.....----.......(((((.(((((((........)))))))..)))))...---------..........)))))))).))..-........... ( -24.51, z-score = -1.77, R) >consensus AUCCACUUUGAUGAA__UUCUUCAAUUCAUGUGUAAUGGUGCGCAUAAUCGAAAAUUUAUGUGCGUCCAUCAG__________AAGGCAAAUGGAACUCAAGUGUUUUUACACUUCC ........(((.((....)).)))...........((((..(((((((........)))))))...))))............................................... ( -9.65 = -9.45 + -0.20)



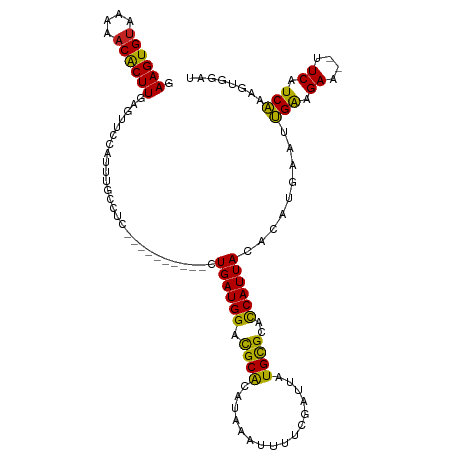
| Location | 5,211,693 – 5,211,797 |
|---|---|
| Length | 104 |
| Sequences | 11 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 79.43 |
| Shannon entropy | 0.40296 |
| G+C content | 0.37646 |
| Mean single sequence MFE | -26.25 |
| Consensus MFE | -13.74 |
| Energy contribution | -12.94 |
| Covariance contribution | -0.81 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.899297 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 5211693 104 + 24543557 GAAGUGUAAAAACACUUGAGUUCCAUUUGCCUC----------CUGAUGGACGCACAUAAAUUUUCGAUUAUGCGCACCAUUAGACAUGAAUUGAAGAA--UUCAUCAAAGUGGAU .((((((....))))))....(((((((.....----------(((((((..((.(((((........))))).)).)))))))..(((((((....))--)))))..))))))). ( -29.40, z-score = -3.58, R) >droSim1.chr3L 4735091 104 + 22553184 GAAGUGUAAAAACACUUGAGUUCCAUUUGCCUC----------CUGAUGGACGCACAUAAAUUUUCGAUUAUGCGCACCAUUAGACAUGAAUUGAAGAA--UUCAUCAAAGUGGAU .((((((....))))))....(((((((.....----------(((((((..((.(((((........))))).)).)))))))..(((((((....))--)))))..))))))). ( -29.40, z-score = -3.58, R) >droSec1.super_2 5165105 104 + 7591821 GAAGUGCAAAAACACUUGAGUUCCAUUUGCCUC----------CUGAUGGACGCAAAUAAAUUUUCGAUUAUGUGCACCAUUAGACAUGAAUUGAAGAA--UUCAUCAAAUUGGAU .(((((......)))))((((((.((((((.((----------(....))).)))))).....(((((((((((..........))))).)))))))))--)))............ ( -25.00, z-score = -2.22, R) >droYak2.chr3L 5774562 104 + 24197627 GAAGUGUAAAAACACUUGAGUUCCAUUUGCCUC----------CUGAUGGACGCACAUAAAUUUUCGAUUAUGCGCACCAUUAGACAUGAAUUGAAGAA--UUCAUCAAAGUGGAU .((((((....))))))....(((((((.....----------(((((((..((.(((((........))))).)).)))))))..(((((((....))--)))))..))))))). ( -29.40, z-score = -3.58, R) >droEre2.scaffold_4784 7906446 104 + 25762168 GAAGUGUAAAAACACUUGAGUUCCAUUUGCCUC----------CUGAUGGACGCACAUAAAUUUUCGAUUAUGCGCACCAUUAGACAUGAAUUGAAGAA--UUCAUCAAAGUGGAU .((((((....))))))....(((((((.....----------(((((((..((.(((((........))))).)).)))))))..(((((((....))--)))))..))))))). ( -29.40, z-score = -3.58, R) >droAna3.scaffold_13337 9967491 104 - 23293914 GAAGUGUAAAGACACUUGAAUUCCAUUUGCCUU----------CUGAUGGACGCGCAUAAAUUUUCGAUUAUGCGCACCAUUACACACGAAUUGAGGAA--UUCAUCAAAGUGGAU .((((((....))))))((((((((((((....----------.((((((..((((((((........)))))))).))))))....)))))...))))--)))............ ( -31.10, z-score = -3.75, R) >dp4.chrXR_group6 7462426 110 + 13314419 GAAGUGUAAAAACACUUGAAUUCCAUUUUGCUUUGCUUUUGUGCUGAUGGACGCACAUAAAUUUUCGAUUAUGCGGACCAUUACACAGGAAUUGAAGAAAAUUCAUCAGA------ .((((((....)))))).((((((.....((...))...((((.((((((.((((..(((........)))))))..))))))))))))))))(((.....)))......------ ( -25.00, z-score = -1.61, R) >droPer1.super_2387 261 110 - 5444 GAAGUGUAAAAACACUUGAAUUCCAUUUUGCUUUGCUUUUGUGCUGAUGGACGCACAUAAAUUUUCGAUUAUGCGGACCAUUACACAGGAAUUGAAGAAAAUUCAUCAGA------ .((((((....)))))).((((((.....((...))...((((.((((((.((((..(((........)))))))..))))))))))))))))(((.....)))......------ ( -25.00, z-score = -1.61, R) >droVir3.scaffold_13049 7945017 100 + 25233164 AAAGUGUAAA-ACGCUUGAAAACCAUUUGGCUUU---------UUGAUGGGCGCACAUAAAUUUGCGAUUAUGCGGUUCAUUACAUAA----CGAAGAGCUCUCAUCGAUAUAA-- .(((((....-.)))))(((((((....)).)))---------))((((((.((.((......))((.(((((..(....)..)))))----))....)).)))))).......-- ( -19.40, z-score = -0.05, R) >droMoj3.scaffold_6680 11602536 102 + 24764193 GAAGUGUAAA-ACGCUUGAGAACCAUUUGGCUGU---------UUGAUGGGCGCACAUAAAUUUGCGAUUAUGCGGUUCAUUACAUAA----CGAAGAGCACUCAUCGAUGUAAAU .(((((....-.)))))..(((((.....(((..---------......)))(((........)))........))))).((((((..----(((.((....)).))))))))).. ( -22.50, z-score = -0.19, R) >droGri2.scaffold_15110 6999776 100 + 24565398 GAAGUGUAAA-ACGCUUGAAAGCCAUUUGACUUU---------UUGAUGGGUGCACAUAAAUUUACGAUUAUGCGGCCCAUUACAUAA----CGGAGAGCUCUCAUCGCUGUAA-- .(((((....-.)))))((.(((..((((.....---------.((((((((.(.(((((........))))).))))))))).....----))))..))).))..........-- ( -23.20, z-score = -0.86, R) >consensus GAAGUGUAAAAACACUUGAGUUCCAUUUGCCUC__________CUGAUGGACGCACAUAAAUUUUCGAUUAUGCGCACCAUUACACAUGAAUUGAAGAA__UUCAUCAAAGUGGAU .((((((....))))))...........................((((((.((((................))))..))))))........((((.((....)).))))....... (-13.74 = -12.94 + -0.81)


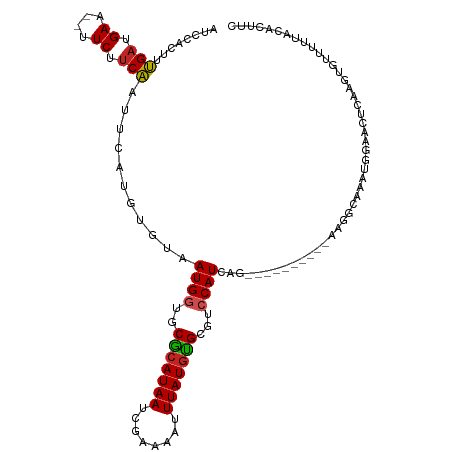
| Location | 5,211,693 – 5,211,797 |
|---|---|
| Length | 104 |
| Sequences | 11 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 79.43 |
| Shannon entropy | 0.40296 |
| G+C content | 0.37646 |
| Mean single sequence MFE | -27.25 |
| Consensus MFE | -9.65 |
| Energy contribution | -9.45 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.776770 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 5211693 104 - 24543557 AUCCACUUUGAUGAA--UUCUUCAAUUCAUGUCUAAUGGUGCGCAUAAUCGAAAAUUUAUGUGCGUCCAUCAG----------GAGGCAAAUGGAACUCAAGUGUUUUUACACUUC .((((.(((((((((--((....))))))).(((.(((((((((((((........))))))))).)))).))----------)...))))))))....((((((....)))))). ( -31.70, z-score = -4.03, R) >droSim1.chr3L 4735091 104 - 22553184 AUCCACUUUGAUGAA--UUCUUCAAUUCAUGUCUAAUGGUGCGCAUAAUCGAAAAUUUAUGUGCGUCCAUCAG----------GAGGCAAAUGGAACUCAAGUGUUUUUACACUUC .((((.(((((((((--((....))))))).(((.(((((((((((((........))))))))).)))).))----------)...))))))))....((((((....)))))). ( -31.70, z-score = -4.03, R) >droSec1.super_2 5165105 104 - 7591821 AUCCAAUUUGAUGAA--UUCUUCAAUUCAUGUCUAAUGGUGCACAUAAUCGAAAAUUUAUUUGCGUCCAUCAG----------GAGGCAAAUGGAACUCAAGUGUUUUUGCACUUC .((((.(((((((((--((....))))))).(((.((((((((.((((........)))).)))).)))).))----------)...))))))))....((((((....)))))). ( -26.40, z-score = -2.29, R) >droYak2.chr3L 5774562 104 - 24197627 AUCCACUUUGAUGAA--UUCUUCAAUUCAUGUCUAAUGGUGCGCAUAAUCGAAAAUUUAUGUGCGUCCAUCAG----------GAGGCAAAUGGAACUCAAGUGUUUUUACACUUC .((((.(((((((((--((....))))))).(((.(((((((((((((........))))))))).)))).))----------)...))))))))....((((((....)))))). ( -31.70, z-score = -4.03, R) >droEre2.scaffold_4784 7906446 104 - 25762168 AUCCACUUUGAUGAA--UUCUUCAAUUCAUGUCUAAUGGUGCGCAUAAUCGAAAAUUUAUGUGCGUCCAUCAG----------GAGGCAAAUGGAACUCAAGUGUUUUUACACUUC .((((.(((((((((--((....))))))).(((.(((((((((((((........))))))))).)))).))----------)...))))))))....((((((....)))))). ( -31.70, z-score = -4.03, R) >droAna3.scaffold_13337 9967491 104 + 23293914 AUCCACUUUGAUGAA--UUCCUCAAUUCGUGUGUAAUGGUGCGCAUAAUCGAAAAUUUAUGCGCGUCCAUCAG----------AAGGCAAAUGGAAUUCAAGUGUCUUUACACUUC ...........((((--((((........(.((..(((((((((((((........))))))))).)))))).----------)........))))))))(((((....))))).. ( -30.59, z-score = -3.41, R) >dp4.chrXR_group6 7462426 110 - 13314419 ------UCUGAUGAAUUUUCUUCAAUUCCUGUGUAAUGGUCCGCAUAAUCGAAAAUUUAUGUGCGUCCAUCAGCACAAAAGCAAAGCAAAAUGGAAUUCAAGUGUUUUUACACUUC ------...((.((....)).))(((((((((((.((((.((((((((........))))))).).))))..)))))...((...)).....)))))).((((((....)))))). ( -26.60, z-score = -2.81, R) >droPer1.super_2387 261 110 + 5444 ------UCUGAUGAAUUUUCUUCAAUUCCUGUGUAAUGGUCCGCAUAAUCGAAAAUUUAUGUGCGUCCAUCAGCACAAAAGCAAAGCAAAAUGGAAUUCAAGUGUUUUUACACUUC ------...((.((....)).))(((((((((((.((((.((((((((........))))))).).))))..)))))...((...)).....)))))).((((((....)))))). ( -26.60, z-score = -2.81, R) >droVir3.scaffold_13049 7945017 100 - 25233164 --UUAUAUCGAUGAGAGCUCUUCG----UUAUGUAAUGAACCGCAUAAUCGCAAAUUUAUGUGCGCCCAUCAA---------AAAGCCAAAUGGUUUUCAAGCGU-UUUACACUUU --.......((((.(.((..((((----((....)))))).(((((((........))))))).)))))))..---------((((((....)))))).......-.......... ( -18.70, z-score = -0.56, R) >droMoj3.scaffold_6680 11602536 102 - 24764193 AUUUACAUCGAUGAGUGCUCUUCG----UUAUGUAAUGAACCGCAUAAUCGCAAAUUUAUGUGCGCCCAUCAA---------ACAGCCAAAUGGUUCUCAAGCGU-UUUACACUUC .........((((.((((..((((----((....))))))..((((((........)))))))))).))))..---------..((((....)))).........-.......... ( -19.60, z-score = -0.70, R) >droGri2.scaffold_15110 6999776 100 - 24565398 --UUACAGCGAUGAGAGCUCUCCG----UUAUGUAAUGGGCCGCAUAAUCGUAAAUUUAUGUGCACCCAUCAA---------AAAGUCAAAUGGCUUUCAAGCGU-UUUACACUUC --.....((..((((((((.....----.......(((((.(((((((........)))))))..)))))...---------..........)))))))).))..-.......... ( -24.51, z-score = -1.91, R) >consensus AUCCACUUUGAUGAA__UUCUUCAAUUCAUGUGUAAUGGUGCGCAUAAUCGAAAAUUUAUGUGCGUCCAUCAG__________AAGGCAAAUGGAACUCAAGUGUUUUUACACUUC ........(((.((....)).)))...........((((..(((((((........)))))))...)))).............................................. ( -9.65 = -9.45 + -0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:04:33 2011