
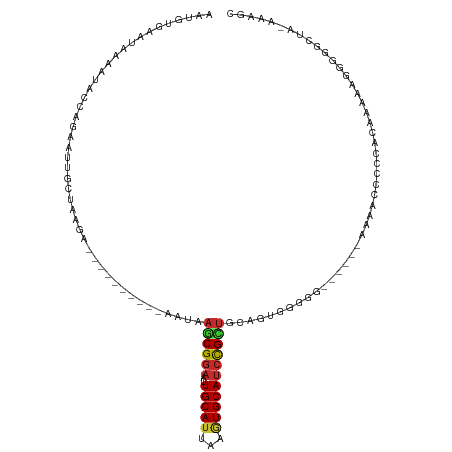
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 5,183,187 – 5,183,287 |
| Length | 100 |
| Max. P | 0.710458 |

| Location | 5,183,187 – 5,183,287 |
|---|---|
| Length | 100 |
| Sequences | 9 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 71.06 |
| Shannon entropy | 0.56079 |
| G+C content | 0.41664 |
| Mean single sequence MFE | -25.07 |
| Consensus MFE | -10.05 |
| Energy contribution | -10.00 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.534483 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
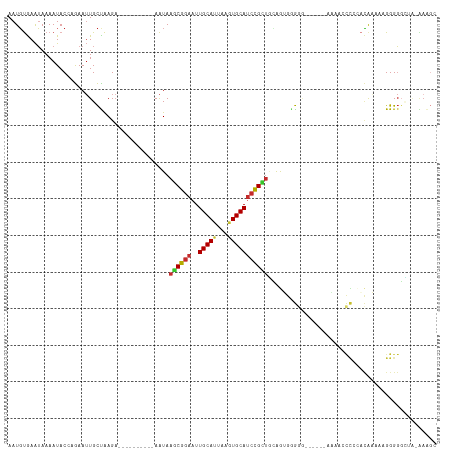
>dm3.chr3L 5183187 100 + 24543557 AAUGUGAAUAAAAUACCUGAAUUGCUAAGA----------AAUAAGCGGAAUUGCAUUAAGUGCAUCCGCUGCAGUGGGGU------AAACCCCCCACAAAAAGGGGGCUA-AAAGC .............(((((..(((((..(..----------..).((((((..(((((...)))))))))))))))).))))------)...(((((.......)))))...-..... ( -31.90, z-score = -2.84, R) >droWil1.scaffold_180698 6877246 102 - 11422946 AAUUCAAGGAAAAUACCAAAAAUGUUGAGCCAAGUAAUGUAGUAAACGGAAUUGCAUUAAAUGCAUCCGCCAAGGACAAGA-------AGA-----AAAUAAAGGGCGUGGUGA--- .............(((((..........((....(((((((((.......)))))))))...))...((((..........-------...-----........))))))))).--- ( -17.70, z-score = -0.37, R) >droPer1.super_153 115004 106 + 195207 AAUGUGAAUAAAAUACAAGAAUUGCCAAGA----------AAUAAACGGAAUUGCAUUAAGUGCAUCCGUUUGAGAAGGAGCGCUGGUUGAAUCGCAUGUCAAGGGGCAGGAGAGG- .....................(((((....----------..((((((((..(((((...))))))))))))).((....(((.(......).)))...))....)))))......- ( -22.50, z-score = -0.58, R) >dp4.chrXR_group6 7421672 106 + 13314419 AAUGUGAAUAAAAUACAAGAAUUGCCAAGA----------AAUAAACGGAAUUGCAUUAAGUGCAUCCGUUUGAGAAGGAGCGCUGGUUGAAUCGCAUGUCAAGGGGCAGGAGAGG- .....................(((((....----------..((((((((..(((((...))))))))))))).((....(((.(......).)))...))....)))))......- ( -22.50, z-score = -0.58, R) >droAna3.scaffold_13337 9938306 89 - 23293914 AAUGUGAAUAAAAUACAAGAAUUGCCAAGA----------AAUAAGCGGAAUUGCAUUAAGUGCAUCCGUUGCGAGGGG---------CAAUCUGUCCAAAAAGGGGG--------- .................(((.(((((....----------....((((((..(((((...)))))))))))......))---------)))))).(((......))).--------- ( -21.14, z-score = -1.34, R) >droEre2.scaffold_4784 7876165 102 + 25762168 AAUGUGAAUAAAAUACCUGAAUUGCUAAGA----------AAUAAGCGGAAUUGCAUUAAGUGCAUCUGCUGUAGUGGGGGGA----AAAACCCCCACAAAAUGGGGGUUA-AAAGC ..............((((..(((.......----------....((((((..(((((...)))))))))))...(((((((..----....)))))))..)))..))))..-..... ( -31.50, z-score = -3.38, R) >droYak2.chr3L 5744899 102 + 24197627 AAUGUGAAUAAAAUACCUGAAUUGCUAAGA----------AAUAAGCGAAAUUGCAUUAAGUGCAGCUGUUGUAGUGGGGGC-----AUAACCUCCACAAAAGGGGGGUUUCGAAGC .......................(((..((----------(((..((((..((((((...))))))...)))).(((((((.-----....))))))).........)))))..))) ( -24.50, z-score = -0.89, R) >droSec1.super_2 5137085 99 + 7591821 AAUGUGAAUAAAAUACCGGAAUUGCUAAGA----------AAUAAGCGGAAUUGCAAUAAGUGCAUCCGCUGCAGUGUGGG------AAAACCCC-ACAAAAGGGGGGCUA-AAAGC ...............(((..(((((..(..----------..).((((((..((((.....))))))))))))))).))).------....((((-......)))).....-..... ( -26.70, z-score = -1.71, R) >droSim1.chr3L 4706093 99 + 22553184 AAUGUGAAUAAAAUACCGGAAUUGCUAAGA----------AAUAAGCGGAAUUGCAUUAAGUGCAUCCGCUGCAGUGUGGG------AAAACCCC-ACAAAAGGGGGGCUA-AAAGC ...............(((..(((((..(..----------..).((((((..(((((...)))))))))))))))).))).------....((((-......)))).....-..... ( -27.20, z-score = -1.73, R) >consensus AAUGUGAAUAAAAUACCAGAAUUGCUAAGA__________AAUAAGCGGAAUUGCAUUAAGUGCAUCCGCUGCAGUGGGGG______AAAACCCCCACAAAAAGGGGGCUA_AAAGC ............................................((((((..(((((...))))))))))).............................................. (-10.05 = -10.00 + -0.05)

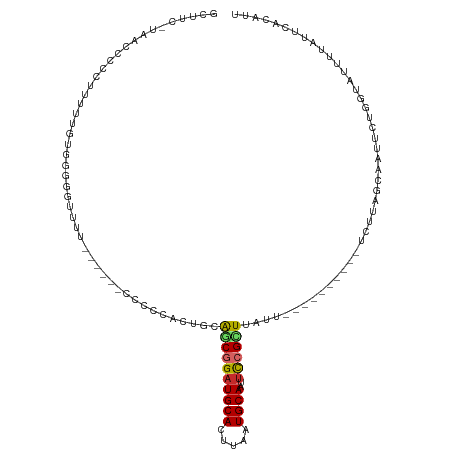
| Location | 5,183,187 – 5,183,287 |
|---|---|
| Length | 100 |
| Sequences | 9 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 71.06 |
| Shannon entropy | 0.56079 |
| G+C content | 0.41664 |
| Mean single sequence MFE | -20.59 |
| Consensus MFE | -8.17 |
| Energy contribution | -8.26 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.710458 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 5183187 100 - 24543557 GCUUU-UAGCCCCCUUUUUGUGGGGGGUUU------ACCCCACUGCAGCGGAUGCACUUAAUGCAAUUCCGCUUAUU----------UCUUAGCAAUUCAGGUAUUUUAUUCACAUU .....-.((((((((......))))))))(------(((....(((((((((((((.....))))..))))))....----------.....))).....))))............. ( -29.20, z-score = -2.54, R) >droWil1.scaffold_180698 6877246 102 + 11422946 ---UCACCACGCCCUUUAUUU-----UCU-------UCUUGUCCUUGGCGGAUGCAUUUAAUGCAAUUCCGUUUACUACAUUACUUGGCUCAACAUUUUUGGUAUUUUCCUUGAAUU ---.......(((........-----...-------...(((....(((((((((((...)))))..))))))....)))......))).....((((..((......))..)))). ( -15.35, z-score = -0.71, R) >droPer1.super_153 115004 106 - 195207 -CCUCUCCUGCCCCUUGACAUGCGAUUCAACCAGCGCUCCUUCUCAAACGGAUGCACUUAAUGCAAUUCCGUUUAUU----------UCUUGGCAAUUCUUGUAUUUUAUUCACAUU -.......((((.........(((..........)))........(((((((((((.....))))..)))))))...----------....))))...................... ( -18.10, z-score = -1.66, R) >dp4.chrXR_group6 7421672 106 - 13314419 -CCUCUCCUGCCCCUUGACAUGCGAUUCAACCAGCGCUCCUUCUCAAACGGAUGCACUUAAUGCAAUUCCGUUUAUU----------UCUUGGCAAUUCUUGUAUUUUAUUCACAUU -.......((((.........(((..........)))........(((((((((((.....))))..)))))))...----------....))))...................... ( -18.10, z-score = -1.66, R) >droAna3.scaffold_13337 9938306 89 + 23293914 ---------CCCCCUUUUUGGACAGAUUG---------CCCCUCGCAACGGAUGCACUUAAUGCAAUUCCGCUUAUU----------UCUUGGCAAUUCUUGUAUUUUAUUCACAUU ---------...((.....))((((((((---------((........((((((((.....))))..))))......----------....)))))))..))).............. ( -16.57, z-score = -1.41, R) >droEre2.scaffold_4784 7876165 102 - 25762168 GCUUU-UAACCCCCAUUUUGUGGGGGUUUU----UCCCCCCACUACAGCAGAUGCACUUAAUGCAAUUCCGCUUAUU----------UCUUAGCAAUUCAGGUAUUUUAUUCACAUU (((..-.............(((((((....----..)))))))...(((.((((((.....))))..)).)))....----------....)))....................... ( -22.40, z-score = -2.24, R) >droYak2.chr3L 5744899 102 - 24197627 GCUUCGAAACCCCCCUUUUGUGGAGGUUAU-----GCCCCCACUACAACAGCUGCACUUAAUGCAAUUUCGCUUAUU----------UCUUAGCAAUUCAGGUAUUUUAUUCACAUU (((..((((..........((((.((....-----.)).))))......(((((((.....)))).....)))..))----------))..)))....................... ( -17.30, z-score = -0.65, R) >droSec1.super_2 5137085 99 - 7591821 GCUUU-UAGCCCCCCUUUUGU-GGGGUUUU------CCCACACUGCAGCGGAUGCACUUAUUGCAAUUCCGCUUAUU----------UCUUAGCAAUUCCGGUAUUUUAUUCACAUU .....-..(((.......(((-(((.....------)))))).(((((((((((((.....))))..))))))....----------.....))).....))).............. ( -24.20, z-score = -2.21, R) >droSim1.chr3L 4706093 99 - 22553184 GCUUU-UAGCCCCCCUUUUGU-GGGGUUUU------CCCACACUGCAGCGGAUGCACUUAAUGCAAUUCCGCUUAUU----------UCUUAGCAAUUCCGGUAUUUUAUUCACAUU .....-..(((.......(((-(((.....------)))))).(((((((((((((.....))))..))))))....----------.....))).....))).............. ( -24.10, z-score = -2.06, R) >consensus GCUUC_UAACCCCCUUUUUGUGGGGGUUUU______CCCCCACUGCAGCGGAUGCACUUAAUGCAAUUCCGCUUAUU__________UCUUAGCAAUUCUGGUAUUUUAUUCACAUU ..............................................((((((((((.....))))..))))))............................................ ( -8.17 = -8.26 + 0.09)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:04:27 2011