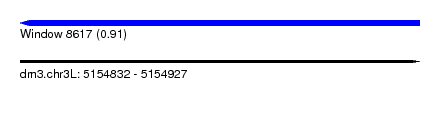
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 5,154,832 – 5,154,927 |
| Length | 95 |
| Max. P | 0.911439 |

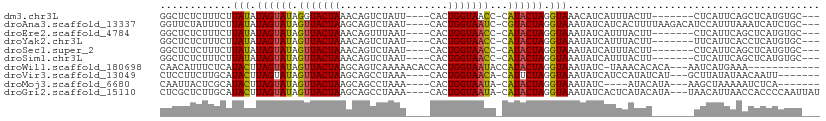
| Location | 5,154,832 – 5,154,927 |
|---|---|
| Length | 95 |
| Sequences | 10 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 72.34 |
| Shannon entropy | 0.55194 |
| G+C content | 0.33362 |
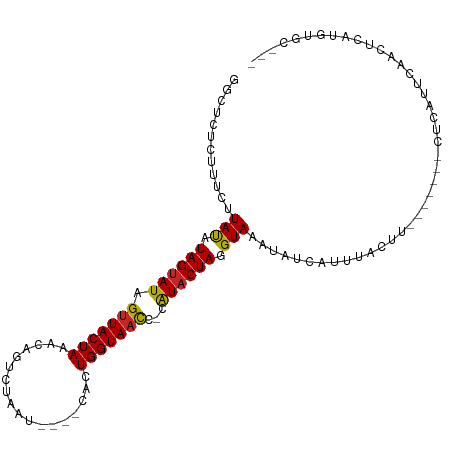
| Mean single sequence MFE | -14.29 |
| Consensus MFE | -8.91 |
| Energy contribution | -8.54 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.911439 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 5154832 95 - 24543557 GGCUCUCUUUCUUAUAUAGUAUAGGUACUAAACAGUCUAUU----CACUGGUAACC-CAUACUAGGUAAACAUCAUUUACUU-------CUCAUUCAGCUCAUGUGC--- ((((............((((((.(((......((((.....----.))))...)))-.))))))((((((.....)))))).-------.......)))).......--- ( -14.90, z-score = -0.85, R) >droAna3.scaffold_13337 9911894 102 + 23293914 GGUUCUAUUUCUUAUAUAGUAUAGUUACUAAGCAGUCUAAU----CACUGGUAAUC-CGUACUAGGUAAAUAUCAUCACUUUUAAGACAUCCAUUUAAAUCAUCUGC--- ((..............(((((....)))))....((((...----..((((((...-..)))))).((((..........))))))))..))...............--- ( -12.00, z-score = 0.18, R) >droEre2.scaffold_4784 7847622 95 - 25762168 GGCUCUCUUUCUUAUAUAGUAUAGUUACUAAACAGUUUAAU----CACUGGUAACC-CAUACUAGGUAAAUAUCAUUUACUU-------CUCAUUCAGCUCAUGUGC--- ((((............((((((.(((((....((((.....----.))))))))).-.))))))(((((((...))))))).-------.......)))).......--- ( -14.90, z-score = -1.22, R) >droYak2.chr3L 5716341 95 - 24197627 GGCUCUCUUUCUUAUAUAGUAUAGUUACUAAACAGUCUAAU----CACUGGUAACC-CAUACUAGGUAAAUAUCAUUUACUU-------UUCAUUCACCUCAUGUGC--- ((..............((((((.(((((....((((.....----.))))))))).-.))))))(((((((...))))))).-------........))........--- ( -13.20, z-score = -0.88, R) >droSec1.super_2 5106804 95 - 7591821 GGCUCUCUUUCUUAUAUAGUAUAGUUACUAAACAGUCUAAU----CACUGGUAACC-CAUACUAGGUAAAUAUCAUUUACUU-------CUCAUUCAGCUCAUGUGC--- ((((............((((((.(((((....((((.....----.))))))))).-.))))))(((((((...))))))).-------.......)))).......--- ( -14.90, z-score = -1.24, R) >droSim1.chr3L 4675013 95 - 22553184 GGCUCUCUUUCUUAUAUAGUAUAGUUACUAAACAGUCUAAU----CACUGGUAACC-CAUACUAGGUAAAUAUCAUUUACUU-------CUCAUUCAGCUCAUGUGC--- ((((............((((((.(((((....((((.....----.))))))))).-.))))))(((((((...))))))).-------.......)))).......--- ( -14.90, z-score = -1.24, R) >droWil1.scaffold_180698 6840622 94 + 11422946 CAACAUUUCUCAUACUUAGUAUAGUUACUAAGCAGUCAAAAACACCACUGGUAAUACCAUACUAGGUAAAUAUC-UAAACACACA---AAUCAUGAAA------------ .........((((.(((((((....)))))))...............((((((......)))))).........-..........---....))))..------------ ( -13.60, z-score = -1.90, R) >droVir3.scaffold_13049 7876176 95 - 25233164 CUCCUUCUUGCAUACUUAGUAUAGUUACUAAGCAGCCUAAA----CACUGGUAACA-CAUUCUAGGUAAAUAUCAUCCAUAUCAU---GCUUAUAUAACAAUU------- .........((((.(((((((....)))))))..(((((..----...((....))-.....)))))................))---)).............------- ( -14.70, z-score = -1.32, R) >droMoj3.scaffold_6680 11528797 91 - 24764193 CAAUUACUCGCAUACUUAGUAUAGUUACUAAGCAGCCUAAA----CACUGGUAAUA-CAUACUAGGUAAAUAUC----AUACAUA---AAGCUAAAAAUCUCA------- ...((((..((...(((((((....)))))))..)).....----..((((((...-..)))))))))).....----.......---...............------- ( -14.40, z-score = -1.66, R) >droGri2.scaffold_15110 6929600 102 - 24565398 CUCGCUCUUGCAUACUUAGUAUAGUUACUAAGCAGCCUAAA----CACUGGUAAUA-CAUACUAGGUAAAUAUCACUCAUACAUA---UAACAUUAACCACCCCAAUUAU ...((....)).((((((((((.(((((((...........----...))))))).-.)))))))))).................---...................... ( -15.44, z-score = -1.41, R) >consensus GGCUCUCUUUCUUAUAUAGUAUAGUUACUAAACAGUCUAAU____CACUGGUAACC_CAUACUAGGUAAAUAUCAUUUACUU_______CUCAUUCAACUCAUGUGC___ ............(((.((((((.(((((((..................)))))))...)))))).))).......................................... ( -8.91 = -8.54 + -0.37)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:04:23 2011