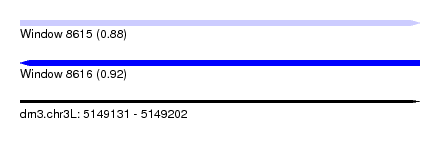
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 5,149,131 – 5,149,202 |
| Length | 71 |
| Max. P | 0.915708 |

| Location | 5,149,131 – 5,149,202 |
|---|---|
| Length | 71 |
| Sequences | 6 |
| Columns | 72 |
| Reading direction | forward |
| Mean pairwise identity | 76.09 |
| Shannon entropy | 0.45770 |
| G+C content | 0.48089 |
| Mean single sequence MFE | -19.47 |
| Consensus MFE | -13.07 |
| Energy contribution | -13.85 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.878702 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
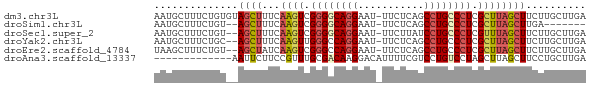
>dm3.chr3L 5149131 71 + 24543557 AAUGCUUUCUGUGUAGCUUUCAAGUCGGGGCAGGAAU-UUCUCAGCCUGCCCUCGCUUAGCUUCUUGCUUGA ..........(((.((((...((((.((((((((...-.......)))))))).))))))))...))).... ( -22.30, z-score = -1.29, R) >droSim1.chr3L 4669391 62 + 22553184 AAUGCUUUCUGU--AGCUUUCAAGUCGGGGCAGGAAU-UUCUCAGCCUGCCCUCGCUUAGCUUGA------- ...(((......--)))..(((((((((((((((...-.......))))))).))....))))))------- ( -21.90, z-score = -1.95, R) >droSec1.super_2 5101161 69 + 7591821 AAUGCUUUCUGU--AGCUUUCAAGUCGGGGCAGGAAU-UUCUUAUCCUGCCCUCGUUUAGCUUCUUGCUUGA ...(((......--)))..((((((((((((((((..-......)))))))).))...........)))))) ( -22.80, z-score = -3.06, R) >droYak2.chr3L 5710576 69 + 24197627 AAUGCUUUCUGC--AGCUUUCAAGUUGGGCCAGGAAU-UUCUCAGCCUGCCCUCGCUUAGCUUCUUGCUUGA ..(((.....))--)(((...((((.((((.(((...-.......)))))))..)))))))........... ( -16.70, z-score = 0.73, R) >droEre2.scaffold_4784 7841939 69 + 25762168 UAAGCUUUCUGU--AGCUAUCAAGUCGGGCCAGGAAU-UUCUCAGCCUGCCCUCGCUUAGCUUCUUGCUUGA (((((.....(.--(((((...(((.((((.(((...-.......)))))))..)))))))).)..))))). ( -19.40, z-score = -0.64, R) >droAna3.scaffold_13337 9906108 59 - 23293914 -------------AAUUCUUCCGUUUGCGACAAGGACAUUUUCGUCCUGUCCUAGCUUAGCUUCCUGCUUGA -------------.........(((.(.((((.((((......))))))))).)))..(((.....)))... ( -13.70, z-score = -2.04, R) >consensus AAUGCUUUCUGU__AGCUUUCAAGUCGGGGCAGGAAU_UUCUCAGCCUGCCCUCGCUUAGCUUCUUGCUUGA ..............((((...((((.((((((((...........)))))))).)))))))).......... (-13.07 = -13.85 + 0.78)
| Location | 5,149,131 – 5,149,202 |
|---|---|
| Length | 71 |
| Sequences | 6 |
| Columns | 72 |
| Reading direction | reverse |
| Mean pairwise identity | 76.09 |
| Shannon entropy | 0.45770 |
| G+C content | 0.48089 |
| Mean single sequence MFE | -19.98 |
| Consensus MFE | -10.57 |
| Energy contribution | -10.85 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.915708 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
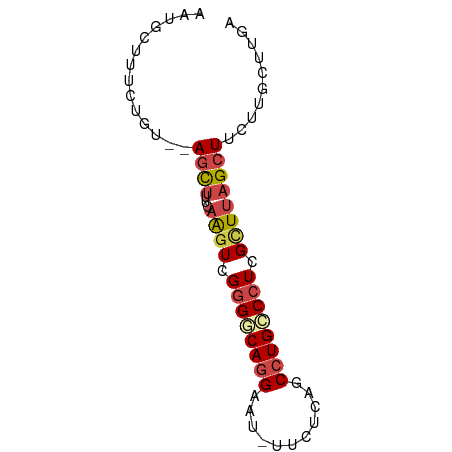
>dm3.chr3L 5149131 71 - 24543557 UCAAGCAAGAAGCUAAGCGAGGGCAGGCUGAGAA-AUUCCUGCCCCGACUUGAAAGCUACACAGAAAGCAUU ...(((.....)))(((((.(((((((..((...-.))))))))))).)))....(((........)))... ( -22.10, z-score = -2.27, R) >droSim1.chr3L 4669391 62 - 22553184 -------UCAAGCUAAGCGAGGGCAGGCUGAGAA-AUUCCUGCCCCGACUUGAAAGCU--ACAGAAAGCAUU -------(((((.....((.(((((((..((...-.))))))))))).)))))..(((--......)))... ( -22.30, z-score = -2.88, R) >droSec1.super_2 5101161 69 - 7591821 UCAAGCAAGAAGCUAAACGAGGGCAGGAUAAGAA-AUUCCUGCCCCGACUUGAAAGCU--ACAGAAAGCAUU ((((((.....)))...((.((((((((......-..))))))))))...)))..(((--......)))... ( -24.00, z-score = -4.70, R) >droYak2.chr3L 5710576 69 - 24197627 UCAAGCAAGAAGCUAAGCGAGGGCAGGCUGAGAA-AUUCCUGGCCCAACUUGAAAGCU--GCAGAAAGCAUU (((((......((...))..(((((((..((...-.))))).))))..)))))..(((--......)))... ( -18.20, z-score = 0.11, R) >droEre2.scaffold_4784 7841939 69 - 25762168 UCAAGCAAGAAGCUAAGCGAGGGCAGGCUGAGAA-AUUCCUGGCCCGACUUGAUAGCU--ACAGAAAGCUUA ..((((..(.(((((((((.((.((((..((...-.)))))).)))).))...)))))--.).....)))). ( -18.80, z-score = -0.35, R) >droAna3.scaffold_13337 9906108 59 + 23293914 UCAAGCAGGAAGCUAAGCUAGGACAGGACGAAAAUGUCCUUGUCGCAAACGGAAGAAUU------------- ....((....(((...)))..((((((((......)))).))))))...(....)....------------- ( -14.50, z-score = -1.73, R) >consensus UCAAGCAAGAAGCUAAGCGAGGGCAGGCUGAGAA_AUUCCUGCCCCGACUUGAAAGCU__ACAGAAAGCAUU ..........((((...((((((((((...........)))))))....)))..)))).............. (-10.57 = -10.85 + 0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:04:22 2011