| Sequence ID | dm3.chr3L |
|---|---|
| Location | 5,123,496 – 5,123,627 |
| Length | 131 |
| Max. P | 0.606889 |

| Location | 5,123,496 – 5,123,599 |
|---|---|
| Length | 103 |
| Sequences | 10 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 60.88 |
| Shannon entropy | 0.84154 |
| G+C content | 0.43383 |
| Mean single sequence MFE | -20.83 |
| Consensus MFE | -7.43 |
| Energy contribution | -7.16 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.86 |
| Mean z-score | -0.79 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.606889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
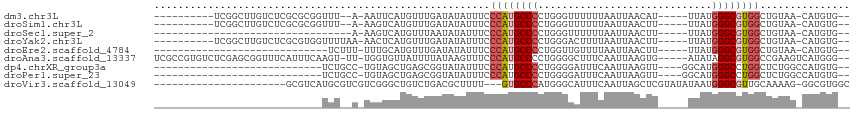
>dm3.chr3L 5123496 103 + 24543557 ---CUCCCACUAAAAAUUCGCCGGGAUUGAGACAUAUCGCACAUG-UUACAGCCACGCCCAUAA-----AUGUUAAUUAAAAAACCCAGGGGCAUGGGAAAUAUAUCAAACA- ---.(((((..............((.(((.(((((.......)))-)).)))))..((((....-----..(((........)))....)))).))))).............- ( -21.20, z-score = -0.64, R) >droSim1.chr3L 4645174 103 + 22553184 ---CUCCCACUAUAAAUUCGCCGGGAUUGAGACAUCUCGCACAUG-UUACAGCCACGCCCAUAA-----AAGUUAAUUAAAAAACCCAGGGGCAUGGGAAAUAUAUCAAACA- ---.(((((..............((.(((.(((((.......)))-)).)))))..((((....-----..(((........)))....)))).))))).............- ( -21.00, z-score = -0.70, R) >droSec1.super_2 5075755 103 + 7591821 ---CUCCCACUAUAAAUUCGCCGGGAUUGAGACAUCUCGCACAUG-UUACAGCCACGCCCAUAA-----AAGUUAAUUAAAAAACCCAGGGGCAUGGGAAAUAUAUUAAACA- ---.(((((..............((.(((.(((((.......)))-)).)))))..((((....-----..(((........)))....)))).))))).............- ( -21.00, z-score = -0.69, R) >droYak2.chr3L 5684750 103 + 24197627 ---CUCCCACUAUAAAUUCGCCGAGAUUGAGACAUCUCACACAUG-UUACAGCCACGCCCAUAA-----AAGUUAAUUAAAAGUCCCAGGGGCAUGGGAAAUAUAUCAAACA- ---.(((((.............(((((......))))).......-..........((((....-----.(.((......)).).....)))).))))).............- ( -18.20, z-score = -0.53, R) >droEre2.scaffold_4784 7816509 96 + 25762168 ----UCCCACUAUAAAUUCAC------UGAGACAUCUCACACAUG-UUACAGCCACGCCCAUAA-----AAGUUAAUUAAAACAACCAGGGGCAUGGGAAAUAUAUCAAACA- ----(((((...........(------((.(((((.......)))-)).)))....((((....-----..(((......)))......)))).))))).............- ( -20.60, z-score = -2.93, R) >droAna3.scaffold_13337 9878119 92 - 23293914 --------------CACCUAC--GAAUGGAGACAUCUCACCCAUGACUUCGGCCACGCCUAUAU-----CACUUAAUUGAAAGCCCCAGGGGCAUGGGAAACUUAUAAAAUAA --------------.......--(((((....))).)).((((((.(((((((...)))....(-----((......)))........))))))))))............... ( -21.40, z-score = -1.47, R) >dp4.chrXR_group3a 1238961 107 + 1468910 CAUCUCCGAAACAGUUUAAACACAAAUUGACACAUCUC-CACAUGGCCAGAGCCAGGCCCAUGCC----AACUUAAUUGAAAUCCCCAGGGGCAUGGGAAAUAUACCGCUCA- .....((....((((((......)))))).........-....((((....))))))((((((((----.......(((.......))).))))))))..............- ( -22.70, z-score = -0.18, R) >droPer1.super_23 1260708 107 + 1662726 CAUCUCCGAAACAGUUUAAACACAAAUUGACACAUCUC-CACAUGGCCAGAGCCAGGCCCAUGCC----AACUUAAUUGAAAUCCCCAGGGGCAUGGGAAAUAUACCGCUCA- .....((....((((((......)))))).........-....((((....))))))((((((((----.......(((.......))).))))))))..............- ( -22.70, z-score = -0.18, R) >droVir3.scaffold_13049 7842528 101 + 25233164 --------GAGCGGCUCAUAAAUAAAUUGCCAGCUCGCGCCACGCCCUUUUGCAACGCCCAUUAUAUACGAGCUAAUUGAAAUGCCCAUGGGCAC---AAAAGCGUCAGACA- --------(((((((.............))).))))((((...((......))...((((((......(((.....)))........))))))..---....))))......- ( -24.36, z-score = -0.24, R) >droGri2.scaffold_15110 17671225 95 - 24565398 ---GCAAUUCAGAAAUCUGAAAUCAGCUGAUGAACUUGAACCAAAAACACAGUUGCGUAUAAAUUAUGCGAGGCAAAUAGAAA--CUGAAAACGUGGACA------------- ---....(((((....))))).(((.....))).......(((......(((((.((((((...))))))...........))--)))......)))...------------- ( -15.10, z-score = -0.28, R) >consensus ___CUCCCACUAAAAAUUCACAGAAAUUGAGACAUCUCACACAUG_UUACAGCCACGCCCAUAA_____AAGUUAAUUAAAAAACCCAGGGGCAUGGGAAAUAUAUCAAACA_ ....................................................(((.((((.............................)))).)))................ ( -7.43 = -7.16 + -0.27)


| Location | 5,123,532 – 5,123,627 |
|---|---|
| Length | 95 |
| Sequences | 9 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 61.27 |
| Shannon entropy | 0.69081 |
| G+C content | 0.45668 |
| Mean single sequence MFE | -24.67 |
| Consensus MFE | -9.59 |
| Energy contribution | -10.05 |
| Covariance contribution | 0.46 |
| Combinations/Pair | 1.12 |
| Mean z-score | -0.99 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.528266 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 5123532 95 - 24543557 ----------UCGGCUUGUCUCGCGCGGUUU--A-AAUUCAUGUUUGAUAUAUUUCCCAUGCCCCUGGGUUUUUUAAUUAACAU-----UUAUGGGCGUGGCUGUAA-CAUGUG-- ----------...((.(((..((.((((...--.-...(((....))).......))(((((((.(((((.((......)).))-----))).)))))))))))..)-)).)).-- ( -21.44, z-score = -0.13, R) >droSim1.chr3L 4645210 95 - 22553184 ----------UCGGCUUGUCUCGCGCGGUUU--A-AAGUCAUGUUUGAUAUAUUUCCCAUGCCCCUGGGUUUUUUAAUUAACUU-----UUAUGGGCGUGGCUGUAA-CAUGUG-- ----------...((.(((..((.((((...--.-..((((....))))......))(((((((..(((((........)))))-----....)))))))))))..)-)).)).-- ( -23.30, z-score = -0.75, R) >droSec1.super_2 5075791 74 - 7591821 ---------------------------------A-AAGUCAUGUUUAAUAUAUUUCCCAUGCCCCUGGGUUUUUUAAUUAACUU-----UUAUGGGCGUGGCUGUAA-CAUGUG-- ---------------------------------.-....(((((....((((....((((((((..(((((........)))))-----....)))))))).)))))-))))..-- ( -19.40, z-score = -2.12, R) >droYak2.chr3L 5684786 97 - 24197627 ----------UCGGCUUGUCUCGCGUGGUUUUAA-AACUCAUGUUUGAUAUAUUUCCCAUGCCCCUGGGACUUUUAAUUAACUU-----UUAUGGGCGUGGCUGUAA-CAUGUG-- ----------.((((.((((..((((((((....-))).)))))..)))).......(((((((.(((((.((......)).))-----))).)))))))))))...-......-- ( -26.10, z-score = -2.08, R) >droEre2.scaffold_4784 7816538 78 - 25762168 -----------------------------UCUUU-UUUGCAUGUUUGAUAUAUUUCCCAUGCCCCUGGUUGUUUUAAUUAACUU-----UUAUGGGCGUGGCUGUAA-CAUGUG-- -----------------------------.....-...(((((((.((......))((((((((.(((..(((......)))..-----))).))))))))....))-))))).-- ( -23.10, z-score = -3.90, R) >droAna3.scaffold_13337 9878142 107 + 23293914 UCGCCGUGUCUCGAGCGGUUUCAUUUCAAGU-UU-UGGUGUUAUUUUAUAAGUUUCCCAUGCCCCUGGGGCUUUCAAUUAAGUG-----AUAUAGGCGUGGCCGAAGUCAUGGG-- ...(((((.((((.(((((..((((......-..-.))))..)))............((((((..((..((((......)))).-----.))..)))))))))).)).))))).-- ( -26.00, z-score = 0.18, R) >dp4.chrXR_group3a 1238999 81 - 1468910 ----------------------------UCUGCC-UGUAGCUGAGCGGUAUAUUUCCCAUGCCCCUGGGGAUUUCAAUUAAGUU----GGCAUGGGCCUGGCUCUGGCCAUGUG-- ----------------------------...(((-.(.((((..((((......))(((((((.((...(((....))).))..----)))))))))..))))).)))......-- ( -23.70, z-score = 1.11, R) >droPer1.super_23 1260746 81 - 1662726 ----------------------------UCUGCC-UGUAGCUGAGCGGUAUAUUUCCCAUGCCCCUGGGGAUUUCAAUUAAGUU----GGCAUGGGCCUGGCUCUGGCCAUGUG-- ----------------------------...(((-.(.((((..((((......))(((((((.((...(((....))).))..----)))))))))..))))).)))......-- ( -23.70, z-score = 1.11, R) >droVir3.scaffold_13049 7842558 90 - 25233164 ----------------------GCGUCAUGCGUCGUCGGGCUGUCUGACGCUUUU---GUGCCCAUGGGCAUUUCAAUUAGCUCGUAUAUAAUGGGCGUUGCAAAAG-GGCGUGGC ----------------------..(((((((..((((((.....))))))(((((---(((((....)))..........((((((.....))))))...)))))))-.))))))) ( -35.30, z-score = -2.32, R) >consensus ____________________________UUU__A_UAGUCAUGUUUGAUAUAUUUCCCAUGCCCCUGGGUAUUUUAAUUAACUU_____UUAUGGGCGUGGCUGUAA_CAUGUG__ ........................................................((((((((.............................))))))))............... ( -9.59 = -10.05 + 0.46)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:04:19 2011