
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 5,119,497 – 5,119,606 |
| Length | 109 |
| Max. P | 0.861016 |

| Location | 5,119,497 – 5,119,606 |
|---|---|
| Length | 109 |
| Sequences | 9 |
| Columns | 125 |
| Reading direction | forward |
| Mean pairwise identity | 75.59 |
| Shannon entropy | 0.46223 |
| G+C content | 0.53563 |
| Mean single sequence MFE | -29.74 |
| Consensus MFE | -21.09 |
| Energy contribution | -21.42 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.861016 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
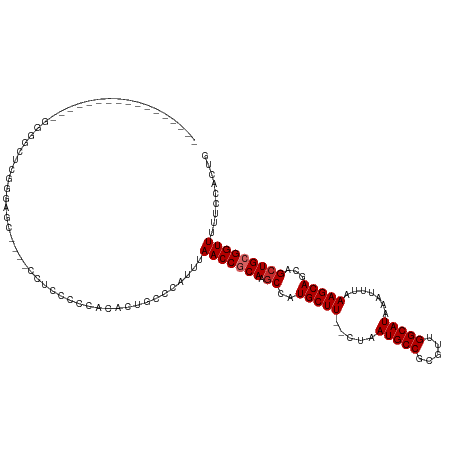
>dm3.chr3L 5119497 109 + 24543557 --------------UGGGGGGUUUGGGUGCUCCCCCCUCCCCUCACUGCCCAUUUAACCGCAAGCCAUGCUU--CUAAUGCCGCGUUGGCAUAAAUUUAAAGCAGCAGCUGCGGUUUUUCCACUG --------------(((((((...(((.....)))...)))))))..........(((((((.((..(((((--...(((((.....))))).......)))))...)))))))))......... ( -38.20, z-score = -1.37, R) >droSim1.chr3L 4641267 102 + 22553184 --------------UUGGGGGUUUGGGU-------UUUCCUCCCACUGCCCAUUUAACCGCAAGCCAUGCUU--CUAAUGCCGCGUUGGCAUAAAUUUAAAGCAGCAGCUGCGGUUUUUCCACUG --------------.(((((((.((((.-------......))))..))))....(((((((.((..(((((--...(((((.....))))).......)))))...)))))))))...)))... ( -36.60, z-score = -2.35, R) >droSec1.super_2 5071896 103 + 7591821 --------------UGGGUGGUUUGGGAG------CUCCCCCAUACUGCCCAUUUAACCGCAAGCCAUGCUU--CUAAUGCCGCGUUGGCAUAAAUUUAAAGCAGCAGCUGCGGUUUUUCCACUG --------------((((..((.((((..------....)))).))..))))...(((((((.((..(((((--...(((((.....))))).......)))))...)))))))))......... ( -40.60, z-score = -3.05, R) >droYak2.chr3L 5680845 92 + 24197627 -------------------------GGAG------CUUCCCCACACUGCCCAUUUAACCGCAAGCCAUGCUU--CUAAUGCCGCGUUGGCAUAAAUUUAAAGCAGCAGCUGCGGUUUUUCCACUG -------------------------((.(------(...........))))....(((((((.((..(((((--...(((((.....))))).......)))))...)))))))))......... ( -25.20, z-score = -1.28, R) >droEre2.scaffold_4784 7812718 101 + 25762168 -----------------GGGGCUCCACCAC----CC-CUCCUCCCCUGCCCAUUUAACCACAAGCCAUGCUU--CUAAUGCCGCGUUGGCAUAAAUUUAAAGCAGCAGCUGCGGUUUUUCCACUG -----------------((((........)----))-).................((((.((.((..(((((--...(((((.....))))).......)))))...)))).))))......... ( -23.10, z-score = -0.14, R) >droAna3.scaffold_13337 9873662 97 - 23293914 -----------------------UGGGUGC---CCAGCCGCCCCCCUGCCCAUUUAACCGCAAGCCAUGCUU--CUAAUGCCGCGUUGGCAUAAAUUUAAAGCAGCAGCUGCGGUUUUUCCACCG -----------------------.(((((.---.....)))))............(((((((.((..(((((--...(((((.....))))).......)))))...)))))))))......... ( -28.50, z-score = -0.59, R) >dp4.chrXR_group3a 1234310 118 + 1468910 CGCCACAGCACCACCCCACCACCCCACCAC----CCACUGCCGUACUACCCAUUUAACCGCAAGCCAUGCUU--CUAAUGCCGCGUUGGCAUAAAUUUAAAGCAGCAGCUGCGGUUUU-CCACUG .((....)).....................----.....................(((((((.((..(((((--...(((((.....))))).......)))))...)))))))))..-...... ( -22.70, z-score = -0.77, R) >droPer1.super_23 1255830 118 + 1662726 CGCCACAGCACCACCCCACCACCCCACCAC----CCACUGCCGUACUACCCAUUUAACCGCAAGCCAUGCUU--CUAAUGCCGCGUUGGCAUAAAUUUAAAGCAGCAGCUGCGGUUUU-CCACUG .((....)).....................----.....................(((((((.((..(((((--...(((((.....))))).......)))))...)))))))))..-...... ( -22.70, z-score = -0.77, R) >droWil1.scaffold_180727 186688 103 + 2741493 ----------------UGGCGGGCGGGAG------UGAACCGAAAGCCUCCCUUUAACCACAAACUAUGCUUUCCUAAUGCCGCGUUGGCAUAAAUUUAAAGCAGCAGCUGCGGUUUUUCCACCG ----------------.((.(((.(((((------.(...(....)))))))...((((.((.....((((((....(((((.....)))))......)))))).....)).))))..))).)). ( -30.10, z-score = -0.43, R) >consensus _________________GGGGCUCGGGAGC____CCUCCCCCACACUGCCCAUUUAACCGCAAGCCAUGCUU__CUAAUGCCGCGUUGGCAUAAAUUUAAAGCAGCAGCUGCGGUUUUUCCACUG .......................................................(((((((.((..(((((.....(((((.....))))).......)))))...)))))))))......... (-21.09 = -21.42 + 0.33)

| Location | 5,119,497 – 5,119,606 |
|---|---|
| Length | 109 |
| Sequences | 9 |
| Columns | 125 |
| Reading direction | reverse |
| Mean pairwise identity | 75.59 |
| Shannon entropy | 0.46223 |
| G+C content | 0.53563 |
| Mean single sequence MFE | -37.20 |
| Consensus MFE | -26.25 |
| Energy contribution | -25.88 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.01 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.763864 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
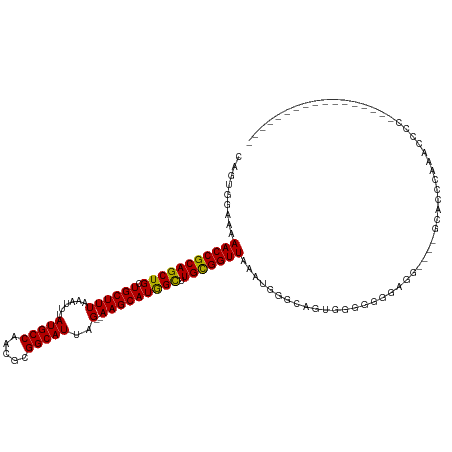
>dm3.chr3L 5119497 109 - 24543557 CAGUGGAAAAACCGCAGCUGCUGCUUUAAAUUUAUGCCAACGCGGCAUUAG--AAGCAUGGCUUGCGGUUAAAUGGGCAGUGAGGGGAGGGGGGAGCACCCAAACCCCCCA-------------- ((.((....(((((((((((.((((((......(((((.....)))))..)--))))))))).))))))).......)).)).((((.((.(((....)))...)))))).-------------- ( -43.10, z-score = -1.51, R) >droSim1.chr3L 4641267 102 - 22553184 CAGUGGAAAAACCGCAGCUGCUGCUUUAAAUUUAUGCCAACGCGGCAUUAG--AAGCAUGGCUUGCGGUUAAAUGGGCAGUGGGAGGAAA-------ACCCAAACCCCCAA-------------- ...(((...(((((((((((.((((((......(((((.....)))))..)--))))))))).)))))))....(((...((((......-------.))))..)))))).-------------- ( -38.60, z-score = -2.44, R) >droSec1.super_2 5071896 103 - 7591821 CAGUGGAAAAACCGCAGCUGCUGCUUUAAAUUUAUGCCAACGCGGCAUUAG--AAGCAUGGCUUGCGGUUAAAUGGGCAGUAUGGGGGAG------CUCCCAAACCACCCA-------------- .........(((((((((((.((((((......(((((.....)))))..)--))))))))).)))))))...((((..((.(((((...------.))))).))..))))-------------- ( -40.20, z-score = -2.21, R) >droYak2.chr3L 5680845 92 - 24197627 CAGUGGAAAAACCGCAGCUGCUGCUUUAAAUUUAUGCCAACGCGGCAUUAG--AAGCAUGGCUUGCGGUUAAAUGGGCAGUGUGGGGAAG------CUCC------------------------- .........(((((((((((.((((((......(((((.....)))))..)--))))))))).)))))))....((((...........)------))).------------------------- ( -29.20, z-score = -0.33, R) >droEre2.scaffold_4784 7812718 101 - 25762168 CAGUGGAAAAACCGCAGCUGCUGCUUUAAAUUUAUGCCAACGCGGCAUUAG--AAGCAUGGCUUGUGGUUAAAUGGGCAGGGGAGGAG-GG----GUGGUGGAGCCCC----------------- .........(((((((((((.((((((......(((((.....)))))..)--))))))))).))))))).................(-((----((......)))))----------------- ( -32.60, z-score = -0.21, R) >droAna3.scaffold_13337 9873662 97 + 23293914 CGGUGGAAAAACCGCAGCUGCUGCUUUAAAUUUAUGCCAACGCGGCAUUAG--AAGCAUGGCUUGCGGUUAAAUGGGCAGGGGGGCGGCUGG---GCACCCA----------------------- .((((......((.(((((((((((((......(((((.....)))))..)--))))....(((((..........)))))..)))))))))---)))))..----------------------- ( -34.70, z-score = -0.14, R) >dp4.chrXR_group3a 1234310 118 - 1468910 CAGUGG-AAAACCGCAGCUGCUGCUUUAAAUUUAUGCCAACGCGGCAUUAG--AAGCAUGGCUUGCGGUUAAAUGGGUAGUACGGCAGUGG----GUGGUGGGGUGGUGGGGUGGUGCUGUGGCG ......-....(((((((.((..(((((.(((((((((.....)))))...--...(((.(((..(.(((..((.....))..))).)..)----)).)))))))..)))))..))))))))).. ( -36.70, z-score = 0.52, R) >droPer1.super_23 1255830 118 - 1662726 CAGUGG-AAAACCGCAGCUGCUGCUUUAAAUUUAUGCCAACGCGGCAUUAG--AAGCAUGGCUUGCGGUUAAAUGGGUAGUACGGCAGUGG----GUGGUGGGGUGGUGGGGUGGUGCUGUGGCG ......-....(((((((.((..(((((.(((((((((.....)))))...--...(((.(((..(.(((..((.....))..))).)..)----)).)))))))..)))))..))))))))).. ( -36.70, z-score = 0.52, R) >droWil1.scaffold_180727 186688 103 - 2741493 CGGUGGAAAAACCGCAGCUGCUGCUUUAAAUUUAUGCCAACGCGGCAUUAGGAAAGCAUAGUUUGUGGUUAAAGGGAGGCUUUCGGUUCA------CUCCCGCCCGCCA---------------- .(((((...(((((((((((.((((((...((.(((((.....))))).)).))))))))).))))))))...(((((............------)))))..))))).---------------- ( -43.00, z-score = -3.28, R) >consensus CAGUGGAAAAACCGCAGCUGCUGCUUUAAAUUUAUGCCAACGCGGCAUUAG__AAGCAUGGCUUGCGGUUAAAUGGGCAGUGGGGGGGAGG____GCACCCAAACCCC_________________ .........(((((((((((.(((((.......(((((.....))))).....))))))))).)))))))....................................................... (-26.25 = -25.88 + -0.37)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:04:18 2011