| Sequence ID | dm3.chr3L |
|---|---|
| Location | 5,101,775 – 5,101,879 |
| Length | 104 |
| Max. P | 0.594625 |

| Location | 5,101,775 – 5,101,879 |
|---|---|
| Length | 104 |
| Sequences | 8 |
| Columns | 132 |
| Reading direction | forward |
| Mean pairwise identity | 68.45 |
| Shannon entropy | 0.52822 |
| G+C content | 0.55892 |
| Mean single sequence MFE | -28.55 |
| Consensus MFE | -13.58 |
| Energy contribution | -14.17 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.594625 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
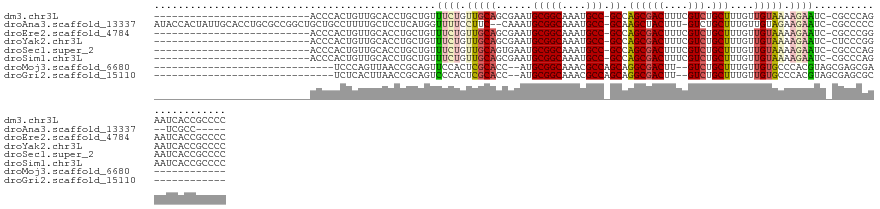
>dm3.chr3L 5101775 104 + 24543557 --------------------------ACCCACUGUUGCACCUGCUGUUUCUGUUGCAGCGAAUGCGGCAAAUGCC-GCCAGCGACUUUCGUCUGCUUUGUUGUAAAAGAAUC-CGCCCAGAAUCACCGCCCC --------------------------.....(((..((....))...((((.((((((((((.(((((....)))-))..(((((....))).)))))))))))).))))..-....)))............ ( -29.00, z-score = -1.74, R) >droAna3.scaffold_13337 9867884 120 - 23293914 AUACCACUAUUGCACCUGCGCCGGCUGCUGCCUUUUGCUCCUCAUGGUUUUCCUUC--CAAAUGCGGCAAAUGCC-GCAAGCUACUUU-GUCUGCUUUGUUGUAGAAGAAUC-CGCCCCC--UCGCC----- .................(((..(((.(((...............(((........)--))..((((((....)))-))))))...(((-.(((((......))))).)))..-.)))...--.))).----- ( -28.40, z-score = -0.55, R) >droEre2.scaffold_4784 7808191 104 + 25762168 --------------------------ACCCACUGUUGCACCUGCUGUUUCUGUUGCAGCGAAUGCGGCAAAUGCC-GCCAGCGACUUUCGUCUGCUUUGUUGUAAAAGAAUC-CGCCCGGAAUCACCGCCCC --------------------------..........((....((.(.((((.((((((((((.(((((....)))-))..(((((....))).)))))))))))).)))).)-.))..((.....))))... ( -31.00, z-score = -1.91, R) >droYak2.chr3L 5675422 104 + 24197627 --------------------------ACCCACUGUUGCACCUGCUGUUUCUGUUGCAGCGAAUGCGGCAAAUGCC-GCCAGCGACUUUCGUCUGCUUUGUUGUAAAAGAAUC-CUCCCGGAAUCACCGCCCC --------------------------..........((.........((((.((((((((((.(((((....)))-))..(((((....))).)))))))))))).))))((-(....)))......))... ( -29.80, z-score = -2.09, R) >droSec1.super_2 5067353 104 + 7591821 --------------------------ACCCACUGUUGCACCUGCUGUUUCUGUUGCAGUGAAUGCGGCAAAUGCC-GCCAGCGACUUUCGUCUGCUUUGUUGUAAAAGAAUC-CGCCCAGAAUCACCGCCCC --------------------------.....(((..((....))...((((.((((((..((.(((((....)))-))..(((((....))).))))..)))))).))))..-....)))............ ( -26.30, z-score = -1.07, R) >droSim1.chr3L 4636463 104 + 22553184 --------------------------ACCCACUGUUGCACCUGCUGUUUCUGUUGCAGCGAAUGCGGCAAAUGCC-GCCAGCGACUUUCGUCUGCUUUGUUGUAAAAGAAUC-CGCCCAGAAUCACCGCCCC --------------------------.....(((..((....))...((((.((((((((((.(((((....)))-))..(((((....))).)))))))))))).))))..-....)))............ ( -29.00, z-score = -1.74, R) >droMoj3.scaffold_6680 11486781 86 + 24764193 ------------------------------UCCCAGUUAACCGCAGUUCCACUCGCACC--AUGCGGCAAACGCCAGCAGGCGACUU--GUCUGCUUUGUUGUGCCCACGUAGCGAGCGA------------ ------------------------------...................(.(((((..(--.((.((((((((..(((((((.....--))))))).)))).)))))).)..))))).).------------ ( -27.80, z-score = -0.83, R) >droGri2.scaffold_15110 6886858 86 + 24565398 ------------------------------UCUCACUUAACCGCAGUCCCACUCGCACC--AUGCGGCAAACGCCAGCAGGCGACUU--GUCUGCUUUGUUGUGCCCACGUAGCGAGCGC------------ ------------------------------...................(.(((((..(--.((.((((((((..(((((((.....--))))))).)))).)))))).)..))))).).------------ ( -27.10, z-score = -0.57, R) >consensus __________________________ACCCACUGUUGCACCUGCUGUUUCUGUUGCAGCGAAUGCGGCAAAUGCC_GCCAGCGACUUUCGUCUGCUUUGUUGUAAAAGAAUC_CGCCCGGAAUCACCGCCCC ...............................................((((.(((((.(((..(((((....))).)).((((((....))).)))))).))))).))))...................... (-13.58 = -14.17 + 0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:04:16 2011