| Sequence ID | dm3.chr3L |
|---|---|
| Location | 5,080,299 – 5,080,407 |
| Length | 108 |
| Max. P | 0.547384 |

| Location | 5,080,299 – 5,080,407 |
|---|---|
| Length | 108 |
| Sequences | 7 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 72.34 |
| Shannon entropy | 0.55661 |
| G+C content | 0.53632 |
| Mean single sequence MFE | -36.12 |
| Consensus MFE | -21.70 |
| Energy contribution | -20.36 |
| Covariance contribution | -1.35 |
| Combinations/Pair | 1.80 |
| Mean z-score | -0.63 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.547384 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 5080299 108 - 24543557 CAGCGAGGCGUCAUCCAAGUGGCCAAAUGUUGUGGUUGUUUGU-GCGGCGUCUGGUUUGCAUUUCC---CUGCAUUAGGAGCUUGGCUGCCACCAGAUAUGCCAGAUGUCCC ......((((((...((((..((((.......))))..)))).-..((((((((((.((((.....---.))))...(((((...))).)))))))))..))).)))))).. ( -37.40, z-score = -0.73, R) >droSim1.chr3L 4615133 108 - 22553184 CAGCGAGGCGUCAUCCAAGUGGCCGAAUGUUGUGGUUGUUUGU-GCGGCGUCUGGUUUGCAUUUCC---CUGCAUUAGGAGCUUGGCUGCCACCAGAUAUGCCAGAUGUCCC ......((((((...((((..((((.......))))..)))).-..((((((((((.((((.....---.))))...(((((...))).)))))))))..))).)))))).. ( -36.70, z-score = -0.44, R) >droSec1.super_2 5046065 108 - 7591821 CAGCGAGGCGUCAUCCAAGUGGCCGAAUGUUGUGGUUGUUUGU-GUGGCGUCUGGUUUGCAUUUCC---CUGCAUUAGGAGCUUGGCUGCCACCAGAUAUGCCAGAGGUCCC .(((.((((((((..((((..((((.......))))..)))).-.)))))))).)))((((.....---.))))...(((.((((((.............)))).)).))). ( -40.52, z-score = -1.59, R) >droYak2.chr3L 5653239 108 - 24197627 CAGCGAGGCGUCAUCCAAGUGGCCGAAUGUUGUGGCUCUUUGU-GCGGCGUCUGGUUUGCAUUUCC---CUGCAUUAGGAGCUUGUGUGCCACCAGAUAUGCCAGAUGUCCC ......((((((...((((.(((((.......))))).)))).-..((((((((((.((((.....---.))))...((.((....)).)))))))))..))).)))))).. ( -34.90, z-score = -0.10, R) >droEre2.scaffold_4784 7783137 108 - 25762168 CAGCGAGGCGUCAUCCAAGUGGCCGAAUGAUGUGGUUGUUGGU-GCGGCGUCUGGUUUGCAUUUCC---CUGCACUAGGAGCUUGGGUGCCACCAGAUAUGCCAGAUGUCCC ......((((((..(((.(..((((.......))))..)))).-..((((((((((..(((...((---(.((.......))..)))))).)))))))..))).)))))).. ( -36.60, z-score = 0.18, R) >droAna3.scaffold_13337 3158708 103 - 23293914 CUGCGAGUCGUCAUCCAAGUGGCCGAAUGUUGCGGUUGUUUGU-GCAACGGAUGUUUUUCAAUGUU---C--CAAUAUGCUCUU---UGAGGAGAUAUAUCAUAGAUGUCAU ......(.((((...((((..((((.......))))..)))).-......((((((((((((.((.---.--......))...)---))))))...)))))...)))).).. ( -22.60, z-score = 0.76, R) >droGri2.scaffold_15110 23900229 100 + 24565398 ---------GUUGACUUAG-GGCCAGUUGUUGCUGCCACAGACGGUGGUGUUUGUUGGGUGCUGCUAGGCUGUCCCAACAAACUAUGUUGCAACAGCUGCAUGAGGCACC-- ---------..((.((((.-.((.(((((((((.(((((.....)))))((((((((((.(((....)))...))))))))))......))))))))))).)))).))..-- ( -44.10, z-score = -2.47, R) >consensus CAGCGAGGCGUCAUCCAAGUGGCCGAAUGUUGUGGUUGUUUGU_GCGGCGUCUGGUUUGCAUUUCC___CUGCAUUAGGAGCUUGGCUGCCACCAGAUAUGCCAGAUGUCCC ......((((((...((((..((((.......))))..))))....((((((((((.((((.........))))......((......)).)))))..))))).)))))).. (-21.70 = -20.36 + -1.35)

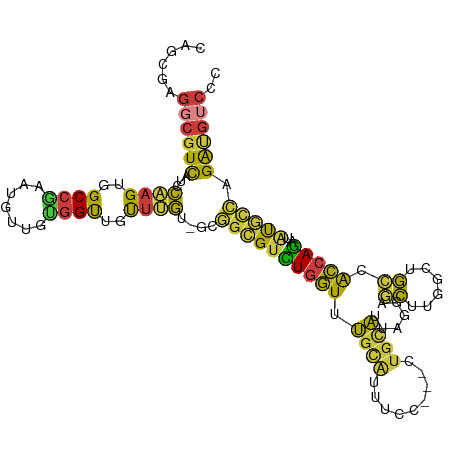
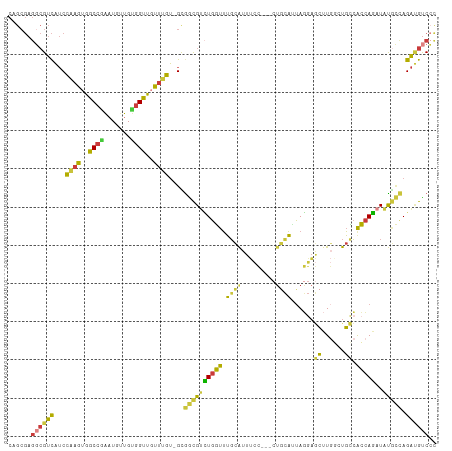
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:04:13 2011