| Sequence ID | dm3.chr3L |
|---|---|
| Location | 5,017,963 – 5,018,055 |
| Length | 92 |
| Max. P | 0.970655 |

| Location | 5,017,963 – 5,018,055 |
|---|---|
| Length | 92 |
| Sequences | 9 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 83.95 |
| Shannon entropy | 0.29974 |
| G+C content | 0.47142 |
| Mean single sequence MFE | -30.34 |
| Consensus MFE | -22.03 |
| Energy contribution | -22.26 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.970655 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
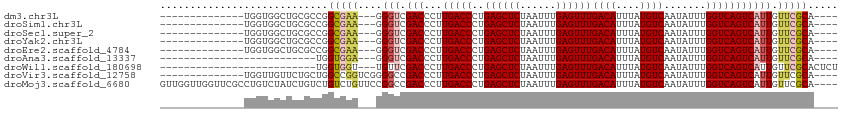
>dm3.chr3L 5017963 92 + 24543557 --------------UGGUGGCUGCGCCGGCGAA---GGGUCGACCCUUGACCCUGAGCUCUAAUUUGAGUUUGACAUUUAUGUCAAUAUUUGGUCAGUCAUUGUUCGCA---- --------------.((((((((.((((((..(---(((((((...))))))))..))).......((((((((((....)))))).))))))))))))))).......---- ( -33.60, z-score = -2.83, R) >droSim1.chr3L 4565407 92 + 22553184 --------------UGGUGGCUGCGCCGGCGAA---GGGUCGACCCUUGACCCUGAGCUCUAAUUUGAGUUUGACAUUUAUGUCAAUAUUUGGUCAGUCAUUGUUCGCA---- --------------.((((((((.((((((..(---(((((((...))))))))..))).......((((((((((....)))))).))))))))))))))).......---- ( -33.60, z-score = -2.83, R) >droSec1.super_2 4997874 92 + 7591821 --------------UGGUGGCUGCGCCGGCGAA---GGGUCGACCCUUGACCCUGAGCUCUAAUUUGAGUUUGACAUUUAUGUCAAUAUUUGGUCAGUCAUUGUUCGCA---- --------------.((((((((.((((((..(---(((((((...))))))))..))).......((((((((((....)))))).))))))))))))))).......---- ( -33.60, z-score = -2.83, R) >droYak2.chr3L 5602666 92 + 24197627 --------------UGGUGGCUGCGCCGGCGAA---GGGUCGACCCUUGACCCUGAGCUCUAAUUUGAGUUUGACAUUUAUGUCAAUAUUUGGUCAGUCAUUGUUCGCA---- --------------.((((((((.((((((..(---(((((((...))))))))..))).......((((((((((....)))))).))))))))))))))).......---- ( -33.60, z-score = -2.83, R) >droEre2.scaffold_4784 7731434 92 + 25762168 --------------UGGUGGCUGCGCCGGCGAA---GGGUCGACCCUUGACCCUGAGCUCUAAUUUGAGUUUGACAUUUAUGUCAAUAUUUGGUCAGUCAUUGUUCGCA---- --------------.((((((((.((((((..(---(((((((...))))))))..))).......((((((((((....)))))).))))))))))))))).......---- ( -33.60, z-score = -2.83, R) >droAna3.scaffold_13337 3110072 80 + 23293914 --------------------------UGGUGGA---GGGUCGACCCUUGACCCUGAGCUCUAAUUUGAGUUUGACAUUUAUGUCAAUAUUUGGUCAGUCAUUGUUCGCA---- --------------------------..(((((---.(((.(((...(((((..((((((......))))))((((....)))).......))))))))))).))))).---- ( -25.10, z-score = -2.16, R) >droWil1.scaffold_180698 1141598 84 - 11422946 --------------------------UGGUGGU---UGUUCGACCCUUGACCCUGAGCUCUAAUUUGAGUUUGACAUUUAUGUCAAUAUUUGGUCAGUCAUUGUUCGCACUCU --------------------------..((((.---.....(((...(((((..((((((......))))))((((....)))).......)))))))).....))))..... ( -20.10, z-score = -1.15, R) >droVir3.scaffold_12758 784166 95 + 972128 --------------UGGUUGUUCUGCUGGCCGGUCGGGGCCGACCCUUGACCCUGAGCUCUAAUUUGAGUUUGACAUUUAUGUCAAUAUUUGGUCAGUCAUUGUUCGCA---- --------------..........(((((((((((((((.....))))))))..((((((......))))))((((....)))).......)))))))...........---- ( -30.90, z-score = -1.93, R) >droMoj3.scaffold_6680 1841503 109 + 24764193 GUUGGUUGGUUCGCCUGUCUAUCUGUCUGUCUGUUCCGGCCGACCCUUGACCCUGAGCUCUAAUUUGAGUUUGACAUUUAUGUCAAUAUUUGGUCAGUCAUUGUUCGCA---- (((((((((..((...(.(.....).)....))..)))))))))..((((((..((((((......))))))((((....)))).......))))))............---- ( -29.00, z-score = -2.22, R) >consensus ______________UGGUGGCUGCGCCGGCGAA___GGGUCGACCCUUGACCCUGAGCUCUAAUUUGAGUUUGACAUUUAUGUCAAUAUUUGGUCAGUCAUUGUUCGCA____ ............................(((((...(((....)))((((((..((((((......))))))((((....)))).......))))))......)))))..... (-22.03 = -22.26 + 0.22)
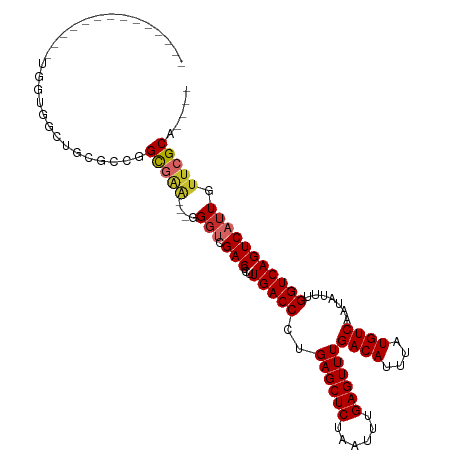
| Location | 5,017,963 – 5,018,055 |
|---|---|
| Length | 92 |
| Sequences | 9 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 83.95 |
| Shannon entropy | 0.29974 |
| G+C content | 0.47142 |
| Mean single sequence MFE | -24.68 |
| Consensus MFE | -16.50 |
| Energy contribution | -16.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.834176 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 5017963 92 - 24543557 ----UGCGAACAAUGACUGACCAAAUAUUGACAUAAAUGUCAAACUCAAAUUAGAGCUCAGGGUCAAGGGUCGACCC---UUCGCCGGCGCAGCCACCA-------------- ----((((........((((.......((((((....)))))).(((......))).))))(((.(((((....)))---)).)))..)))).......-------------- ( -27.00, z-score = -2.38, R) >droSim1.chr3L 4565407 92 - 22553184 ----UGCGAACAAUGACUGACCAAAUAUUGACAUAAAUGUCAAACUCAAAUUAGAGCUCAGGGUCAAGGGUCGACCC---UUCGCCGGCGCAGCCACCA-------------- ----((((........((((.......((((((....)))))).(((......))).))))(((.(((((....)))---)).)))..)))).......-------------- ( -27.00, z-score = -2.38, R) >droSec1.super_2 4997874 92 - 7591821 ----UGCGAACAAUGACUGACCAAAUAUUGACAUAAAUGUCAAACUCAAAUUAGAGCUCAGGGUCAAGGGUCGACCC---UUCGCCGGCGCAGCCACCA-------------- ----((((........((((.......((((((....)))))).(((......))).))))(((.(((((....)))---)).)))..)))).......-------------- ( -27.00, z-score = -2.38, R) >droYak2.chr3L 5602666 92 - 24197627 ----UGCGAACAAUGACUGACCAAAUAUUGACAUAAAUGUCAAACUCAAAUUAGAGCUCAGGGUCAAGGGUCGACCC---UUCGCCGGCGCAGCCACCA-------------- ----((((........((((.......((((((....)))))).(((......))).))))(((.(((((....)))---)).)))..)))).......-------------- ( -27.00, z-score = -2.38, R) >droEre2.scaffold_4784 7731434 92 - 25762168 ----UGCGAACAAUGACUGACCAAAUAUUGACAUAAAUGUCAAACUCAAAUUAGAGCUCAGGGUCAAGGGUCGACCC---UUCGCCGGCGCAGCCACCA-------------- ----((((........((((.......((((((....)))))).(((......))).))))(((.(((((....)))---)).)))..)))).......-------------- ( -27.00, z-score = -2.38, R) >droAna3.scaffold_13337 3110072 80 - 23293914 ----UGCGAACAAUGACUGACCAAAUAUUGACAUAAAUGUCAAACUCAAAUUAGAGCUCAGGGUCAAGGGUCGACCC---UCCACCA-------------------------- ----............((((.......((((((....)))))).(((......))).))))(((..((((....)))---)..))).-------------------------- ( -20.30, z-score = -2.14, R) >droWil1.scaffold_180698 1141598 84 + 11422946 AGAGUGCGAACAAUGACUGACCAAAUAUUGACAUAAAUGUCAAACUCAAAUUAGAGCUCAGGGUCAAGGGUCGAACA---ACCACCA-------------------------- .(.(((.(.....(((((((((.....((((((....)))))).(((......))).....))))...)))))....---.))))).-------------------------- ( -17.90, z-score = -1.16, R) >droVir3.scaffold_12758 784166 95 - 972128 ----UGCGAACAAUGACUGACCAAAUAUUGACAUAAAUGUCAAACUCAAAUUAGAGCUCAGGGUCAAGGGUCGGCCCCGACCGGCCAGCAGAACAACCA-------------- ----(((..........(((((.....((((((....)))))).(((......))).....)))))..((((((......)))))).))).........-------------- ( -27.50, z-score = -2.49, R) >droMoj3.scaffold_6680 1841503 109 - 24764193 ----UGCGAACAAUGACUGACCAAAUAUUGACAUAAAUGUCAAACUCAAAUUAGAGCUCAGGGUCAAGGGUCGGCCGGAACAGACAGACAGAUAGACAGGCGAACCAACCAAC ----.............(((((.....((((((....)))))).(((......))).....)))))..(((..(((......................)))..)))....... ( -21.45, z-score = -0.67, R) >consensus ____UGCGAACAAUGACUGACCAAAUAUUGACAUAAAUGUCAAACUCAAAUUAGAGCUCAGGGUCAAGGGUCGACCC___UUCGCCGGCGCAGCCACCA______________ .............(((((((((.....((((((....)))))).(((......))).....))))...)))))........................................ (-16.50 = -16.50 + 0.00)

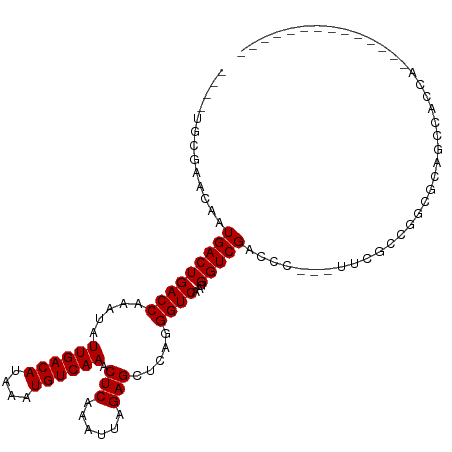
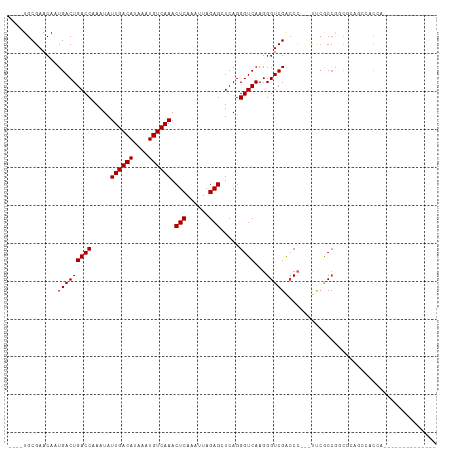
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:04:08 2011