
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 5,014,821 – 5,014,942 |
| Length | 121 |
| Max. P | 0.753626 |

| Location | 5,014,821 – 5,014,925 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 67.01 |
| Shannon entropy | 0.50196 |
| G+C content | 0.36156 |
| Mean single sequence MFE | -16.21 |
| Consensus MFE | -9.10 |
| Energy contribution | -8.91 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.753626 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
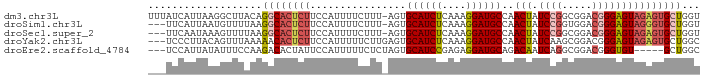
>dm3.chr3L 5014821 104 + 24543557 UCGAAUAUUUUUCCAAAAUGAUUUCUUUAUUUUAUCAUUAAGGCUUACAGGCACUCUUCCAUUUU-CUUUAGUGCAUCUCAAAGGAUGCCAACUAUCCGGCGGAC ...........(((..((((((...........))))))...(((.....(((((..........-....)))))........(((((.....))))))))))). ( -17.04, z-score = -0.54, R) >droSim1.chr3L 4562318 90 + 22553184 -------------UCAAAU-AUUUUAAAAAUAUUUCAUUAAUGUUUUAAGGCACUCUUCCAUUUU-CUUUAGUGCAUCUCAAAGGAUGCCAACUAUCCGGUGGAC -------------......-......((((((((.....))))))))..........((((((..-...((((((((((....))))))..))))...)))))). ( -16.70, z-score = -1.94, R) >droYak2.chr3L 5599507 87 + 24197627 ------------------UGAAAUAUUUCUAACUCCCUUACAGUUUAAAAACACUCUUCCAUUUUUCUUGAGUGCAUCUCAAAGGAUGCCAACUAUCAAGCGGAC ------------------...............((((((..((((.......((((.............))))((((((....)))))).))))...))).))). ( -14.02, z-score = -1.22, R) >droEre2.scaffold_4784 7728314 87 + 25762168 ------------------UCAAAUAUUUUGUACUCCAUUAUAUUUCCAAGACACUAUUCCAUUUUUCUCUAGUGCAUCCGAGAGGAUGCAGACAAUCAGGCGGAC ------------------..((((((..((.....))..))))))............(((............(((((((....)))))))(.....)....))). ( -17.10, z-score = -1.46, R) >consensus __________________UGAAAUAUUUAUUACUCCAUUAAAGUUUAAAGACACUCUUCCAUUUU_CUUUAGUGCAUCUCAAAGGAUGCCAACUAUCAGGCGGAC .........................................................(((.............((((((....))))))............))). ( -9.10 = -8.91 + -0.19)


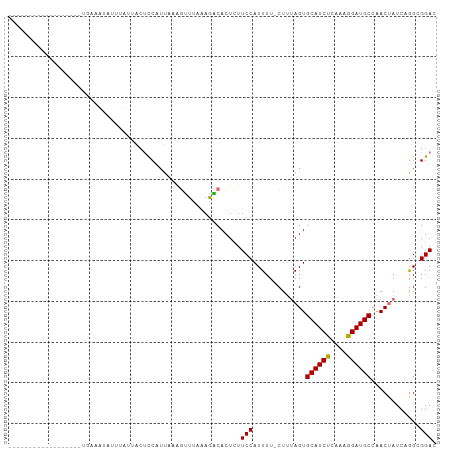
| Location | 5,014,851 – 5,014,942 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 80.42 |
| Shannon entropy | 0.34350 |
| G+C content | 0.45471 |
| Mean single sequence MFE | -24.78 |
| Consensus MFE | -17.50 |
| Energy contribution | -18.50 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.680105 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
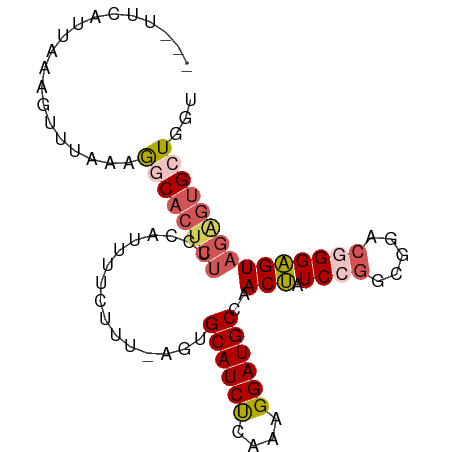
>dm3.chr3L 5014851 91 + 24543557 UUUAUCAUUAAGGCUUACAGGCACUCUUCCAUUUUCUUU-AGUGCAUCUCAAAGGAUGCCAACUAUCCGGCGGACGGGAGUAGAGUGCUGGU ................((.((((((((............-.(.((((((....))))))).(((.((((.....))))))))))))))).)) ( -25.80, z-score = -1.36, R) >droSim1.chr3L 4562337 88 + 22553184 ---UUCAUUAAUGUUUUAAGGCACUCUUCCAUUUUCUUU-AGUGCAUCUCAAAGGAUGCCAACUAUCCGGUGGACGGGAGUAGGGUGCUGGU ---................((((((((............-.(.((((((....))))))).(((.((((.....)))))))))))))))... ( -24.60, z-score = -1.25, R) >droSec1.super_2 4994830 88 + 7591821 ---UUCAAUAAAGUUUUAAGGCACUCUUCCAUUUUCUUU-AGUGCAUCUCAAAGGAUGCCAACUAUCCGGCGGACGGGAGUAGAGUGCUGGU ---................((((((((............-.(.((((((....))))))).(((.((((.....)))))))))))))))... ( -25.50, z-score = -1.81, R) >droYak2.chr3L 5599522 89 + 24197627 ---UCCCUUACAGUUUAAAAACACUCUUCCAUUUUUCUUGAGUGCAUCUCAAAGGAUGCCAACUAUCAAGCGGACGGGAGUAGAGUGCUGGC ---.......(((........(((((((((..((..(((((..((((((....))))))......)))))..))..)))..))))))))).. ( -22.90, z-score = -1.02, R) >droEre2.scaffold_4784 7728329 84 + 25762168 ---UCCAUUAUAUUUCCAAGACACUAUUCCAUUUUUCUCUAGUGCAUCCGAGAGGAUGCAGACAAUCAGGCGGACGGGUGU-----GCUGGC ---............(((..(((((..(((............(((((((....)))))))(.....)....)))..)))))-----..))). ( -25.10, z-score = -1.85, R) >consensus ___UUCAUUAAAGUUUAAAGGCACUCUUCCAUUUUCUUU_AGUGCAUCUCAAAGGAUGCCAACUAUCCGGCGGACGGGAGUAGAGUGCUGGU ...................(((((((((((.((..((...(((((((((....))))))..)))....))..)).)))...))))))))... (-17.50 = -18.50 + 1.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:04:07 2011