| Sequence ID | dm3.chr3L |
|---|---|
| Location | 5,008,887 – 5,008,980 |
| Length | 93 |
| Max. P | 0.998715 |

| Location | 5,008,887 – 5,008,980 |
|---|---|
| Length | 93 |
| Sequences | 10 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 91.23 |
| Shannon entropy | 0.19124 |
| G+C content | 0.42340 |
| Mean single sequence MFE | -32.83 |
| Consensus MFE | -26.51 |
| Energy contribution | -26.69 |
| Covariance contribution | 0.18 |
| Combinations/Pair | 1.24 |
| Mean z-score | -4.10 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.46 |
| SVM RNA-class probability | 0.998715 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
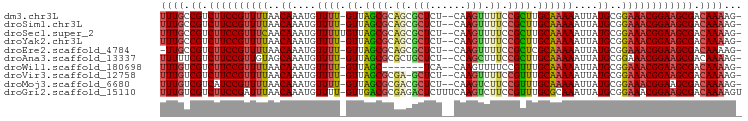
>dm3.chr3L 5008887 93 + 24543557 UUUGCCGUCUUCCGUUUUAACAAAUGUUUU-GUUAGCGCAGCGCUCU--CAAGUUUUCCGCUUGCAAAAAUUAUGCGGAAACGGAAGCGACAAAAG- ((((.((.((((((((((..((....((((-((.((((.((.(((..--..))).)).)))).))))))....))..)))))))))))).))))..- ( -31.40, z-score = -3.38, R) >droSim1.chr3L 4556291 93 + 22553184 UUUGCCGUCUUCCGUUUUAACAAAUGUUUU-GUUAGCGCAGCGCUCU--CAAGUUUUCCGCUUGCAAAAAUUAUGCGGAAACGGAAGCGACAAAAG- ((((.((.((((((((((..((....((((-((.((((.((.(((..--..))).)).)))).))))))....))..)))))))))))).))))..- ( -31.40, z-score = -3.38, R) >droSec1.super_2 4988763 94 + 7591821 UUUGCCGUCUUCCGUUUCAACAAAUGUUUUUGUUAGCGCAGCGCUCU--CAAGUUUUCCGCUUGCAAAAAUUAUGCGGAAACGGAAGCGACAAAAG- ((((.((.((((((((((..((...((((((((.((((.((.(((..--..))).)).)))).))))))))..))..)))))))))))).))))..- ( -35.90, z-score = -4.80, R) >droYak2.chr3L 5593354 93 + 24197627 UUUGCCGUCUUCCGUUUUAACAAAUGUUUU-GUUAGCGCAGCGCUCU--CAAGUUUUCCGCUUGCAAAAAUUAUGCGGAAACGGAAGCGACAAAAG- ((((.((.((((((((((..((....((((-((.((((.((.(((..--..))).)).)))).))))))....))..)))))))))))).))))..- ( -31.40, z-score = -3.38, R) >droEre2.scaffold_4784 7722467 92 + 25762168 -UUGCCGUCUUCCGUUUUAACAAAUGUUUU-GUUAGCGCAGCGCUCU--CAAGUUUUCCGCUCGCAAAAAUUAUGCGGAAACGGAAGCGACAAAAG- -(((.((.((((((((((..((....((((-((.((((.((.(((..--..))).)).)))).))))))....))..)))))))))))).)))...- ( -30.50, z-score = -3.14, R) >droAna3.scaffold_13337 3101776 93 + 23293914 UUUUUCGUCUUCCGUUGUAGCAAAUGUUUU-GUUAGCGCGCUGCUCU--CCAGCUUUCCGCUUGCAAAAAUUAUGCGGAAACGGAAGCGACAAAAG- (((.(((.((((((((...(((....((((-((.((((.((((....--.))))....)))).))))))....)))...))))))))))).)))..- ( -34.30, z-score = -3.94, R) >droWil1.scaffold_180698 1131584 86 - 11422946 UUUGUCGUCUUCCGUUUUAACAAAUGUUUU-GUUAGC-------UCA--CAAGUUUUCCGUUUGCAAAAAUUAUGCGGAAACGGAAGCGACAAAAG- (((((((.((((((((((..((....((((-((.(((-------...--..........))).))))))....))..)))))))))))))))))..- ( -29.22, z-score = -4.76, R) >droVir3.scaffold_12758 769848 92 + 972128 UUUGUCGUCUUCCGUUUUAACAAAUGUUUU-GUUAGCGCGA-GCUCU--CAAGUUUUCCGUUUGCAAAAAUUAUGCGGAAACGGAAGCGACAAAAG- (((((((.((((((((((..((....((((-((.((((.((-(((..--..)))))..)))).))))))....))..)))))))))))))))))..- ( -35.40, z-score = -5.36, R) >droMoj3.scaffold_6680 1831303 93 + 24764193 UUUGUCGUCAUCCGUUUUAACAAAUGUUUU-GUUAGCGCGACGCUCU--CAAGUCUUCCGUUUGCAAAAAUUAUGCGGAAACGGAAGCGACAAAAG- ((((((((..((((((((..((....((((-((.((((.(((.....--...)))...)))).))))))....))..)))))))).))))))))..- ( -31.80, z-score = -4.05, R) >droGri2.scaffold_15110 23834861 96 - 24565398 UUUGUCGUCUUCCGAUUUAACAAAUGUUUU-GUUGACGCGAGACUCUUUCAAGUCUUCCGUUUGCGCAAAUUAUGCGGAAACGGAAGCGACAAAAGU (((((((((((.((..(((((((.....))-))))))).)))))........(.(((((((((.((((.....)))).))))))))))))))))... ( -37.00, z-score = -4.80, R) >consensus UUUGCCGUCUUCCGUUUUAACAAAUGUUUU_GUUAGCGCAGCGCUCU__CAAGUUUUCCGCUUGCAAAAAUUAUGCGGAAACGGAAGCGACAAAAG_ ((((.((.((((((((((..((...(((((.((.((((.((.(((......))).)).)))).)).)))))..))..)))))))))))).))))... (-26.51 = -26.69 + 0.18)

| Location | 5,008,887 – 5,008,980 |
|---|---|
| Length | 93 |
| Sequences | 10 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 91.23 |
| Shannon entropy | 0.19124 |
| G+C content | 0.42340 |
| Mean single sequence MFE | -30.86 |
| Consensus MFE | -24.13 |
| Energy contribution | -24.19 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.41 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.78 |
| SVM RNA-class probability | 0.995192 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
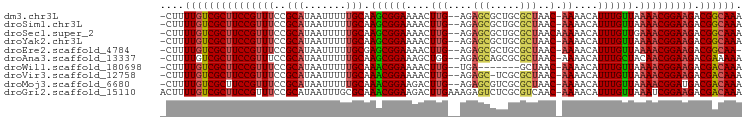
>dm3.chr3L 5008887 93 - 24543557 -CUUUUGUCGCUUCCGUUUCCGCAUAAUUUUUGCAAGCGGAAAACUUG--AGAGCGCUGCGCUAAC-AAAACAUUUGUUAAAACGGAAGACGGCAAA -..((((((((((((((((((((.............)))))....(((--..((((...))))..)-))............)))))))).))))))) ( -30.32, z-score = -2.76, R) >droSim1.chr3L 4556291 93 - 22553184 -CUUUUGUCGCUUCCGUUUCCGCAUAAUUUUUGCAAGCGGAAAACUUG--AGAGCGCUGCGCUAAC-AAAACAUUUGUUAAAACGGAAGACGGCAAA -..((((((((((((((((((((.............)))))....(((--..((((...))))..)-))............)))))))).))))))) ( -30.32, z-score = -2.76, R) >droSec1.super_2 4988763 94 - 7591821 -CUUUUGUCGCUUCCGUUUCCGCAUAAUUUUUGCAAGCGGAAAACUUG--AGAGCGCUGCGCUAACAAAAACAUUUGUUGAAACGGAAGACGGCAAA -..(((((((((((((((((.(((...((((((..((((.(...((..--..))...).))))..))))))....))).)))))))))).))))))) ( -34.20, z-score = -3.73, R) >droYak2.chr3L 5593354 93 - 24197627 -CUUUUGUCGCUUCCGUUUCCGCAUAAUUUUUGCAAGCGGAAAACUUG--AGAGCGCUGCGCUAAC-AAAACAUUUGUUAAAACGGAAGACGGCAAA -..((((((((((((((((((((.............)))))....(((--..((((...))))..)-))............)))))))).))))))) ( -30.32, z-score = -2.76, R) >droEre2.scaffold_4784 7722467 92 - 25762168 -CUUUUGUCGCUUCCGUUUCCGCAUAAUUUUUGCGAGCGGAAAACUUG--AGAGCGCUGCGCUAAC-AAAACAUUUGUUAAAACGGAAGACGGCAA- -...(((((((((((((((.((((.......))))((((((....(((--..((((...))))..)-))....)))))).))))))))).))))))- ( -30.90, z-score = -2.69, R) >droAna3.scaffold_13337 3101776 93 - 23293914 -CUUUUGUCGCUUCCGUUUCCGCAUAAUUUUUGCAAGCGGAAAGCUGG--AGAGCAGCGCGCUAAC-AAAACAUUUGCUACAACGGAAGACGAAAAA -......(((((((((((...(((....(((((..((((....((((.--....)))).))))..)-))))....)))...)))))))).))).... ( -30.90, z-score = -2.68, R) >droWil1.scaffold_180698 1131584 86 + 11422946 -CUUUUGUCGCUUCCGUUUCCGCAUAAUUUUUGCAAACGGAAAACUUG--UGA-------GCUAAC-AAAACAUUUGUUAAAACGGAAGACGACAAA -..((((((((((((((((.((((...((((((....))))))...))--)).-------..((((-((.....))))))))))))))).))))))) ( -28.30, z-score = -4.75, R) >droVir3.scaffold_12758 769848 92 - 972128 -CUUUUGUCGCUUCCGUUUCCGCAUAAUUUUUGCAAACGGAAAACUUG--AGAGC-UCGCGCUAAC-AAAACAUUUGUUAAAACGGAAGACGACAAA -..((((((((((((((((..(((.......))).((((((....(((--..(((-....)))..)-))....)))))).))))))))).))))))) ( -29.60, z-score = -4.13, R) >droMoj3.scaffold_6680 1831303 93 - 24764193 -CUUUUGUCGCUUCCGUUUCCGCAUAAUUUUUGCAAACGGAAGACUUG--AGAGCGUCGCGCUAAC-AAAACAUUUGUUAAAACGGAUGACGACAAA -..((((((((((((((((..(((.......))))))))))))..(((--..((((...))))..)-))..(((((((....))))))).))))))) ( -29.70, z-score = -3.38, R) >droGri2.scaffold_15110 23834861 96 + 24565398 ACUUUUGUCGCUUCCGUUUCCGCAUAAUUUGCGCAAACGGAAGACUUGAAAGAGUCUCGCGUCAAC-AAAACAUUUGUUAAAUCGGAAGACGACAAA ....(((.(((((((((((.((((.....)))).))))))))(((((....)))))..))).))).-......((((((...((....)).)))))) ( -34.00, z-score = -4.43, R) >consensus _CUUUUGUCGCUUCCGUUUCCGCAUAAUUUUUGCAAGCGGAAAACUUG__AGAGCGCUGCGCUAAC_AAAACAUUUGUUAAAACGGAAGACGACAAA ....(((((((((((((((..(((.......))).((((((...........(((.....)))..........)))))).))))))))).)))))). (-24.13 = -24.19 + 0.06)
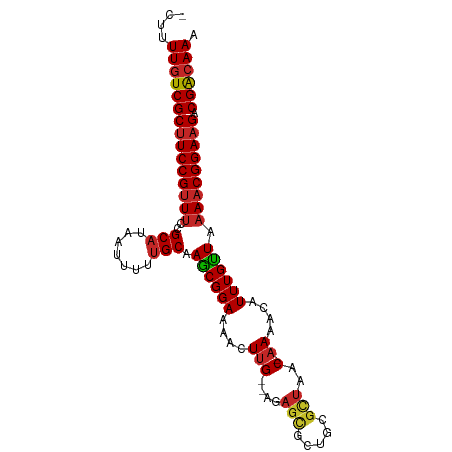
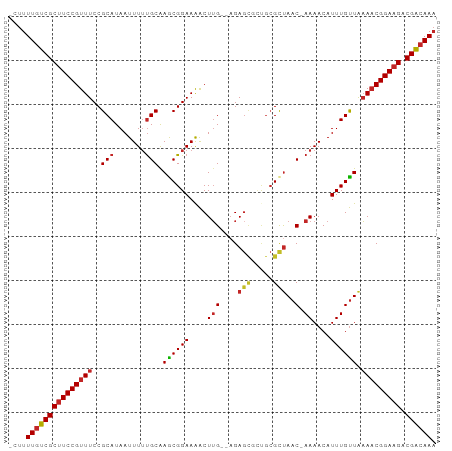
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:04:05 2011