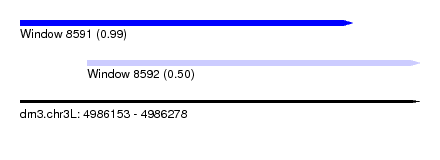
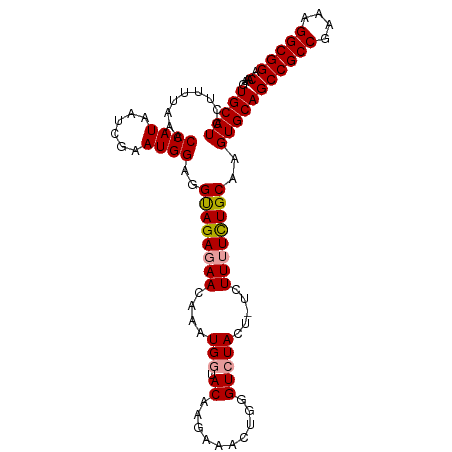
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 4,986,153 – 4,986,278 |
| Length | 125 |
| Max. P | 0.985073 |

| Location | 4,986,153 – 4,986,257 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 91.32 |
| Shannon entropy | 0.12120 |
| G+C content | 0.43876 |
| Mean single sequence MFE | -32.07 |
| Consensus MFE | -25.20 |
| Energy contribution | -25.09 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.20 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.985073 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 4986153 104 + 24543557 UGCAUCCUUUUAAACCAUAAUCGAAUGGAGGUAGAGAACAAAUGGUACAAGAAAAUGGGUCUACUACUUUUUUUGCAAGUGCAGCCGCCGAAAGGCGGACAAAG (((((.((((....((((......))))....))))...........(((((((((((.....))).))))))))...)))))((((((....))))).).... ( -28.50, z-score = -2.25, R) >droSec1.super_2 4966216 103 + 7591821 UGCAUACUUUUAAACCAUAAUCGAAUGGAAGUAGAUAACAAAUGCUACCAGAAACUGGGUCUACU-UCUUUUCUGCAAGUGCAGCCGCCGAAAGGCGGACAAAG (((((....(((.....)))..(((.((((((((((..(....)...((((...)))))))))))-))).))).....)))))((((((....))))).).... ( -32.60, z-score = -4.04, R) >droSim1.chr3L 4533200 103 + 22553184 UGCAUCCUUUUAAACCAUAAUCGGAUGGAGGCAGAGAACAAAUGGUACAAGAAACUGGGUCUACU-UGUUUUCUGCAAGUGCAGCCGCCGAAUGGCGGACAAAG (((((.........((((......))))..(((((((((((.(((..((......))...))).)-))))))))))..)))))((((((....))))).).... ( -35.10, z-score = -3.31, R) >consensus UGCAUCCUUUUAAACCAUAAUCGAAUGGAGGUAGAGAACAAAUGGUACAAGAAACUGGGUCUACU_UCUUUUCUGCAAGUGCAGCCGCCGAAAGGCGGACAAAG (((((.........((((......))))..((((((((........((..........))........))))))))..)))))((((((....))))).).... (-25.20 = -25.09 + -0.11)

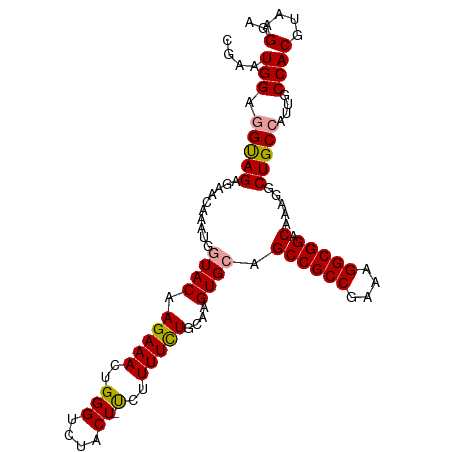
| Location | 4,986,174 – 4,986,278 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 91.32 |
| Shannon entropy | 0.12120 |
| G+C content | 0.50006 |
| Mean single sequence MFE | -35.00 |
| Consensus MFE | -24.14 |
| Energy contribution | -24.03 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chr3L 4986174 104 + 24543557 CGAAUGGAGGUAGAGAACAAAUGGUACAAGAAAAUGGGUCUACUACUUUUUUUGCAAGUGCAGCCGCCGAAAGGCGGACAAAGCCUGCCAUUGCCACGUAAGGA ..(((((((((............((((((((((((((.....))).)))))))....)))).((((((....))))).)...)))).))))).((......)). ( -30.60, z-score = -1.47, R) >droSec1.super_2 4966237 103 + 7591821 CGAAUGGAAGUAGAUAACAAAUGCUACCAGAAACUGGGUCUACU-UCUUUUCUGCAAGUGCAGCCGCCGAAAGGCGGACAAAGGCUGCCAUUGCCACGUAAGGA .(((.((((((((((..(....)...((((...)))))))))))-))).))).((((..(((((((((....))).......))))))..))))..(....).. ( -38.51, z-score = -3.52, R) >droSim1.chr3L 4533221 103 + 22553184 CGGAUGGAGGCAGAGAACAAAUGGUACAAGAAACUGGGUCUACU-UGUUUUCUGCAAGUGCAGCCGCCGAAUGGCGGACAAAGGCUGCCAUUGCCACGUAAGGA ....(((..(((((((((((.(((..((......))...))).)-))))))))))..(.((((((.(((.....))).....)))))))....)))(....).. ( -35.90, z-score = -1.99, R) >consensus CGAAUGGAGGUAGAGAACAAAUGGUACAAGAAACUGGGUCUACU_UCUUUUCUGCAAGUGCAGCCGCCGAAAGGCGGACAAAGGCUGCCAUUGCCACGUAAGGA ....(((.(((((......................................((((....))))(((((....))))).......)))))....)))........ (-24.14 = -24.03 + -0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:04:02 2011