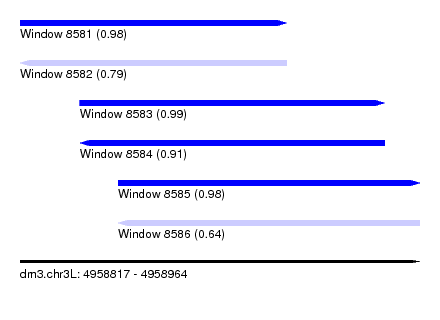
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 4,958,817 – 4,958,964 |
| Length | 147 |
| Max. P | 0.987350 |

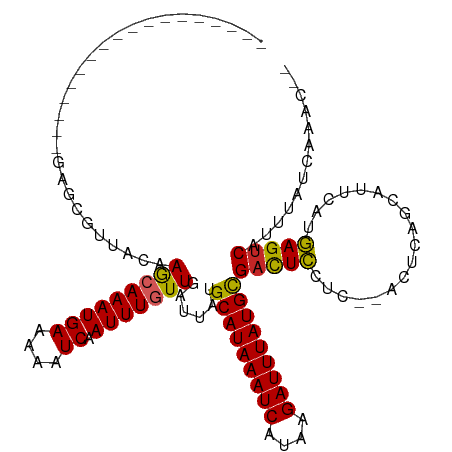
| Location | 4,958,817 – 4,958,915 |
|---|---|
| Length | 98 |
| Sequences | 10 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.74 |
| Shannon entropy | 0.27754 |
| G+C content | 0.31202 |
| Mean single sequence MFE | -22.17 |
| Consensus MFE | -17.40 |
| Energy contribution | -16.95 |
| Covariance contribution | -0.45 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.984760 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
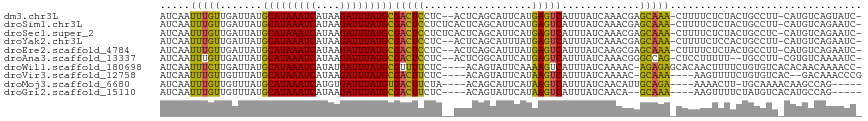
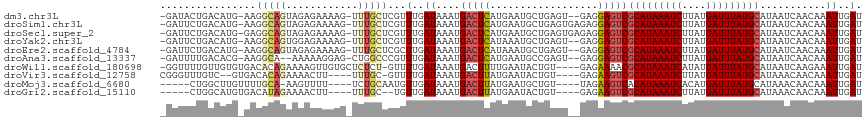
>dm3.chr3L 4958817 98 + 24543557 ------------------GAGCGUUACAAGCAAAUGAAAAAUCAAUUUGUUGAUUAUGCAUAAAUCAUAAGAUUUAUGCGACUCCUC--ACUCAGCAUUCAUGAGUCAUUUAUCAAAC-- ------------------((((.......))((((((....(((...((((((....(((((((((....)))))))))((....))--..))))))....))).)))))).))....-- ( -23.30, z-score = -3.08, R) >droSim1.chr3L 4497084 100 + 22553184 ------------------GAGCGUUACAAGCAAAUGAAAAAUCAAUUUGUUGAUUAUGCAUAAAUCAUAAGAUUUAUGCGACUCCUCUCACUCAGCAUUCAUGAGUCAUUUAUCAAAC-- ------------------((((.......))((((((....(((...((((((....(((((((((....)))))))))((......))..))))))....))).)))))).))....-- ( -23.50, z-score = -3.01, R) >droSec1.super_2 4939272 100 + 7591821 ------------------GAGCGUUACAAGCAAAUGAAAAAUCAAUUUGUUGAUUAUGCAUAAAUCAUAAGAUUUAUGCGACUCCUCUCACUCAGCAUUCAUGAGUCAUUUAUCAAAC-- ------------------((((.......))((((((....(((...((((((....(((((((((....)))))))))((......))..))))))....))).)))))).))....-- ( -23.50, z-score = -3.01, R) >droYak2.chr3L 5541358 98 + 24197627 ------------------GAGCGUUACAAACAAAUGAAAAAUCAAUUUGUUGAUUAUGCAUAAAUCAUAAGAUUUAUGCGACUCCUC--ACUCAGCAUUUAUGAGUCAUUUAUCAAAC-- ------------------(((.(((...(((((((((....)).)))))))......(((((((((....))))))))))))..)))--(((((.......)))))............-- ( -22.40, z-score = -3.33, R) >droEre2.scaffold_4784 7670911 98 + 25762168 ------------------GAGCGUUACAAGCAAAUGAAAAAUCAAUUUGUUGAUUAUGCAUAAAUCAUAAGAUUUAUGCGACUCCUC--ACUCAGCAUUUAUGAGUCAUUUAUCAAGC-- ------------------((((.......))((((((....(((...((((((....(((((((((....)))))))))((....))--..))))))....))).)))))).))....-- ( -23.30, z-score = -2.69, R) >droAna3.scaffold_13337 3050117 98 + 23293914 ------------------GAGCAUUACAAGAAAAUGAAAAAUCAAUUUGUUGAUUAUGCAUAAAUCAUAAGAUUUAUGCGACUCCUC--ACUCGGCAUUCAUGAGUCAUUUAUCAAAC-- ------------------...........((((((((....(((...((((((....(((((((((....)))))))))((....))--..))))))....))).)))))).))....-- ( -21.70, z-score = -2.61, R) >droWil1.scaffold_180698 1068390 114 - 11422946 ACAAAAACGAGGCGUGGCAAGCAUUACAAGCAAAUGAAAAAUCAAUUUCUUGAUUAUGCAUAAAUCAUAUGAUUUAUGCGUUUUCUC----ACAGUAUUCAAAAGUCAUUUAUCAAAA-- ........((...(((((..((.......))...((((.((((((....))))))(((((((((((....)))))))))))......----......))))...)))))...))....-- ( -24.40, z-score = -2.68, R) >droVir3.scaffold_12758 698862 97 + 972128 ------------------AGCGAUUGCAAGCAAAUGAAAAAUCAAUUUGUUGUUUAUGCAUAAAUCAUAAGAUUUAUGCGACUUCUC----ACAGUAUUCAUAAGUCAUUUAUCAAAAC- ------------------...((.(((.(((((((((....)).))))))).....((((((((((....)))))))))).......----...))).))...................- ( -19.10, z-score = -1.70, R) >droMoj3.scaffold_6680 1767930 98 + 24764193 ------------------AGGGCGUGUCAGCAAAUGAAAAAUCAAUUUGUUGUUUAUGCAUAAAUCAUGUGAUUUAUGUGACUUCUA----ACAGCAUUCAUAAGUCAUUUAUCAACAUU ------------------...(((((.((((((((((....)).))))))))..)))))(((((((....)))))))(((((((...----...........)))))))........... ( -20.94, z-score = -1.40, R) >droGri2.scaffold_15110 23770965 96 - 24565398 ------------------GAUCAGUGCAAGCAAAUGAAAAAUCAAUUUGUUGUUUAUGCAUAAAUCAUAAGAUUUAUGCGACUUCUC----ACAGUAUUCAUAAGUCAUUUAUCAACA-- ------------------(((.(((((.(((((((((....)).))))))).....((((((((((....)))))))))).......----.............).)))).)))....-- ( -19.60, z-score = -2.11, R) >consensus __________________GAGCGUUACAAGCAAAUGAAAAAUCAAUUUGUUGAUUAUGCAUAAAUCAUAAGAUUUAUGCGACUCCUC__ACUCAGCAUUCAUGAGUCAUUUAUCAAAC__ ............................(((((((((....)).)))))))......(((((((((....)))))))))(((((..................)))))............. (-17.40 = -16.95 + -0.45)

| Location | 4,958,817 – 4,958,915 |
|---|---|
| Length | 98 |
| Sequences | 10 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.74 |
| Shannon entropy | 0.27754 |
| G+C content | 0.31202 |
| Mean single sequence MFE | -22.14 |
| Consensus MFE | -16.82 |
| Energy contribution | -16.64 |
| Covariance contribution | -0.18 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.790785 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 4958817 98 - 24543557 --GUUUGAUAAAUGACUCAUGAAUGCUGAGU--GAGGAGUCGCAUAAAUCUUAUGAUUUAUGCAUAAUCAACAAAUUGAUUUUUCAUUUGCUUGUAACGCUC------------------ --......................((.((((--(((((((((((((((((....)))))))))..(((......)))))))))))))))))...........------------------ ( -22.70, z-score = -1.57, R) >droSim1.chr3L 4497084 100 - 22553184 --GUUUGAUAAAUGACUCAUGAAUGCUGAGUGAGAGGAGUCGCAUAAAUCUUAUGAUUUAUGCAUAAUCAACAAAUUGAUUUUUCAUUUGCUUGUAACGCUC------------------ --..(((((....(((((....((.....)).....)))))(((((((((....)))))))))...)))))(((((.((....)))))))............------------------ ( -22.30, z-score = -1.08, R) >droSec1.super_2 4939272 100 - 7591821 --GUUUGAUAAAUGACUCAUGAAUGCUGAGUGAGAGGAGUCGCAUAAAUCUUAUGAUUUAUGCAUAAUCAACAAAUUGAUUUUUCAUUUGCUUGUAACGCUC------------------ --..(((((....(((((....((.....)).....)))))(((((((((....)))))))))...)))))(((((.((....)))))))............------------------ ( -22.30, z-score = -1.08, R) >droYak2.chr3L 5541358 98 - 24197627 --GUUUGAUAAAUGACUCAUAAAUGCUGAGU--GAGGAGUCGCAUAAAUCUUAUGAUUUAUGCAUAAUCAACAAAUUGAUUUUUCAUUUGUUUGUAACGCUC------------------ --(((.((((((((((((((.........))--))).....(((((((((....)))))))))..((((((....))))))..)))))))))...)))....------------------ ( -25.60, z-score = -3.01, R) >droEre2.scaffold_4784 7670911 98 - 25762168 --GCUUGAUAAAUGACUCAUAAAUGCUGAGU--GAGGAGUCGCAUAAAUCUUAUGAUUUAUGCAUAAUCAACAAAUUGAUUUUUCAUUUGCUUGUAACGCUC------------------ --(((((((....(((((....((.....))--...)))))(((((((((....)))))))))...)))))(((((.((....)))))))........))..------------------ ( -23.90, z-score = -2.24, R) >droAna3.scaffold_13337 3050117 98 - 23293914 --GUUUGAUAAAUGACUCAUGAAUGCCGAGU--GAGGAGUCGCAUAAAUCUUAUGAUUUAUGCAUAAUCAACAAAUUGAUUUUUCAUUUUCUUGUAAUGCUC------------------ --...((((....(((((....((.....))--...)))))(((((((((....)))))))))...))))((((..(((....))).....)))).......------------------ ( -21.10, z-score = -1.28, R) >droWil1.scaffold_180698 1068390 114 + 11422946 --UUUUGAUAAAUGACUUUUGAAUACUGU----GAGAAAACGCAUAAAUCAUAUGAUUUAUGCAUAAUCAAGAAAUUGAUUUUUCAUUUGCUUGUAAUGCUUGCCACGCCUCGUUUUUGU --.......((((((............((----((((((..(((((((((....)))))))))....((((....))))))))))))..((..(.....)..))......)))))).... ( -23.60, z-score = -1.68, R) >droVir3.scaffold_12758 698862 97 - 972128 -GUUUUGAUAAAUGACUUAUGAAUACUGU----GAGAAGUCGCAUAAAUCUUAUGAUUUAUGCAUAAACAACAAAUUGAUUUUUCAUUUGCUUGCAAUCGCU------------------ -((.((.((((.....)))).)).))((.----.((.....(((((((((....)))))))))........(((((.((....)))))))))..))......------------------ ( -20.30, z-score = -1.67, R) >droMoj3.scaffold_6680 1767930 98 - 24764193 AAUGUUGAUAAAUGACUUAUGAAUGCUGU----UAGAAGUCACAUAAAUCACAUGAUUUAUGCAUAAACAACAAAUUGAUUUUUCAUUUGCUGACACGCCCU------------------ ..((((..(((((((...((.(((..(((----(........((((((((....)))))))).......)))).))).))...)))))))..))))......------------------ ( -19.06, z-score = -1.70, R) >droGri2.scaffold_15110 23770965 96 + 24565398 --UGUUGAUAAAUGACUUAUGAAUACUGU----GAGAAGUCGCAUAAAUCUUAUGAUUUAUGCAUAAACAACAAAUUGAUUUUUCAUUUGCUUGCACUGAUC------------------ --((((.((((.....)))).)))).((.----.((.....(((((((((....)))))))))........(((((.((....)))))))))..))......------------------ ( -20.50, z-score = -1.98, R) >consensus __GUUUGAUAAAUGACUCAUGAAUGCUGAGU__GAGGAGUCGCAUAAAUCUUAUGAUUUAUGCAUAAUCAACAAAUUGAUUUUUCAUUUGCUUGUAACGCUC__________________ .....(((.(((((((((..................)))))(((((((((....)))))))))...............)))).))).................................. (-16.82 = -16.64 + -0.18)

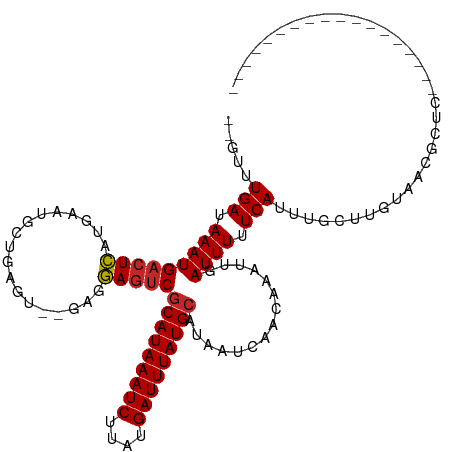
| Location | 4,958,839 – 4,958,951 |
|---|---|
| Length | 112 |
| Sequences | 10 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 79.74 |
| Shannon entropy | 0.41527 |
| G+C content | 0.33689 |
| Mean single sequence MFE | -21.60 |
| Consensus MFE | -16.02 |
| Energy contribution | -15.04 |
| Covariance contribution | -0.98 |
| Combinations/Pair | 1.37 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.27 |
| SVM RNA-class probability | 0.987350 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 4958839 112 + 24543557 AUCAAUUUGUUGAUUAUGCAUAAAUCAUAAGAUUUAUGCGACUCCUC--ACUCAGCAUUCAUGAGUCAUUUAUCAAACGAGCAAA-CUUUUCUCUACUGCCUU-CAUGUCAGUAUC- .....(((((((((...(((((((((....)))))))))(((((...--.............)))))....))).))))))....-........(((((.(..-...).)))))..- ( -23.29, z-score = -2.49, R) >droSim1.chr3L 4497106 114 + 22553184 AUCAAUUUGUUGAUUAUGCAUAAAUCAUAAGAUUUAUGCGACUCCUCUCACUCAGCAUUCAUGAGUCAUUUAUCAAACGAGCAAA-CUUUUCUCUACUGCCUU-CAUGUCAGAAUC- .....(((((((((...(((((((((....)))))))))(((((..................)))))....))).))))))....-..........(((.(..-...).)))....- ( -21.07, z-score = -1.49, R) >droSec1.super_2 4939294 114 + 7591821 AUCAAUUUGUUGAUUAUGCAUAAAUCAUAAGAUUUAUGCGACUCCUCUCACUCAGCAUUCAUGAGUCAUUUAUCAAACGAGCAAA-CUUUUCUCUACUGCCUC-CAUGUCAGAAUC- .....(((((((((...(((((((((....)))))))))(((((..................)))))....))).))))))....-..........(((.(..-...).)))....- ( -21.07, z-score = -1.68, R) >droYak2.chr3L 5541380 112 + 24197627 AUCAAUUUGUUGAUUAUGCAUAAAUCAUAAGAUUUAUGCGACUCCUC--ACUCAGCAUUUAUGAGUCAUUUAUCAAACGAGCAAA-CUUUUCUCCACUGCCUU-CAUGUCAGAAUC- .....(((((((((...(((((((((....)))))))))(((((...--.............)))))....))).))))))....-..........(((.(..-...).)))....- ( -21.19, z-score = -1.74, R) >droEre2.scaffold_4784 7670933 112 + 25762168 AUCAAUUUGUUGAUUAUGCAUAAAUCAUAAGAUUUAUGCGACUCCUC--ACUCAGCAUUUAUGAGUCAUUUAUCAAGCGAGCAAA-CUUUUCUCUACUGCCUU-CAUGUCAGAAUC- .....(((((((((...(((((((((....)))))))))(((((...--.............)))))....))).))))))....-..........(((.(..-...).)))....- ( -21.19, z-score = -1.12, R) >droAna3.scaffold_13337 3050139 110 + 23293914 AUCAAUUUGUUGAUUAUGCAUAAAUCAUAAGAUUUAUGCGACUCCUC--ACUCGGCAUUCAUGAGUCAUUUAUCAAACGGGCCAG-CUCCUUUUU--UGCCUU-CGUGUCAAAAUC- .........((((...((((((((((....))))))))))......(--((..((((....(((........)))...(((....-)))......--))))..-.)))))))....- ( -22.00, z-score = -1.29, R) >droWil1.scaffold_180698 1068430 111 - 11422946 AUCAAUUUCUUGAUUAUGCAUAAAUCAUAUGAUUUAUGCGUUUUCUC----ACAGUAUUCAAAAGUCAUUUAUCAAAAC-AGAGAGCACAACUUUUCUGUGUCACACAACAAAACC- ...........((..(((((((((((....)))))))))))..)).(----((((.....((((((......((.....-.)).......)))))))))))...............- ( -21.62, z-score = -2.90, R) >droVir3.scaffold_12758 698884 106 + 972128 AUCAAUUUGUUGUUUAUGCAUAAAUCAUAAGAUUUAUGCGACUUCUC----ACAGUAUUCAUAAGUCAUUUAUCAAAAC-GCAAA----AAGUUUUCUGUGUCAC--GACAAACCCG .....((((((((...((((((((((....))))))))))......(----((((...................(((((-.....----..))))))))))..))--)))))).... ( -22.00, z-score = -3.28, R) >droMoj3.scaffold_6680 1767952 103 + 24764193 AUCAAUUUGUUGUUUAUGCAUAAAUCAUGUGAUUUAUGUGACUUCUA----ACAGCAUUCAUAAGUCAUUUAUCAACAUUGCAGA----AAAACUU-UGCAAAACAAGCCAG----- .......(((((.....(((((((((....)))))))))(((((...----...........)))))......)))))(((((((----.....))-)))))..........----- ( -20.94, z-score = -1.58, R) >droGri2.scaffold_15110 23770987 102 - 24565398 AUCAAUUUGUUGUUUAUGCAUAAAUCAUAAGAUUUAUGCGACUUCUC----ACAGUAUUCAUAAGUCAUUUAUCAACA--GCAAA----AAGUUUUCUAUGUCACAUGCCAG----- .....(((((((((...(((((((((....)))))))))(((((...----...........))))).......))))--)))))----.......................----- ( -21.64, z-score = -3.62, R) >consensus AUCAAUUUGUUGAUUAUGCAUAAAUCAUAAGAUUUAUGCGACUCCUC__ACUCAGCAUUCAUGAGUCAUUUAUCAAACGAGCAAA_CUUUUCUCUACUGCCUU_CAUGUCAGAAUC_ .....(((((.......(((((((((....)))))))))(((((..................))))).............)))))................................ (-16.02 = -15.04 + -0.98)
| Location | 4,958,839 – 4,958,951 |
|---|---|
| Length | 112 |
| Sequences | 10 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 79.74 |
| Shannon entropy | 0.41527 |
| G+C content | 0.33689 |
| Mean single sequence MFE | -27.17 |
| Consensus MFE | -14.81 |
| Energy contribution | -14.63 |
| Covariance contribution | -0.18 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.912692 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
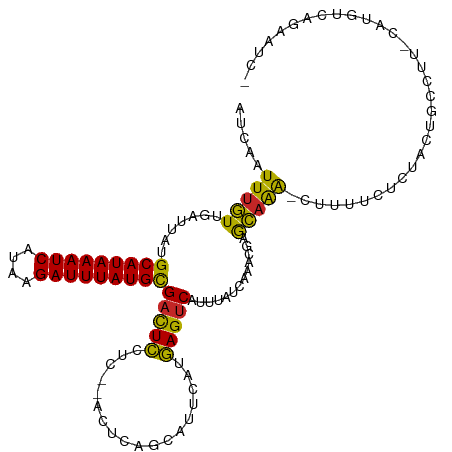
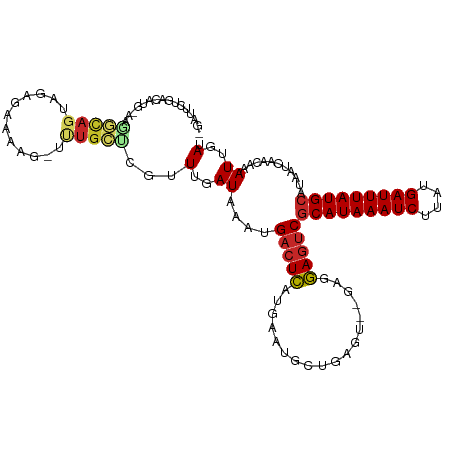
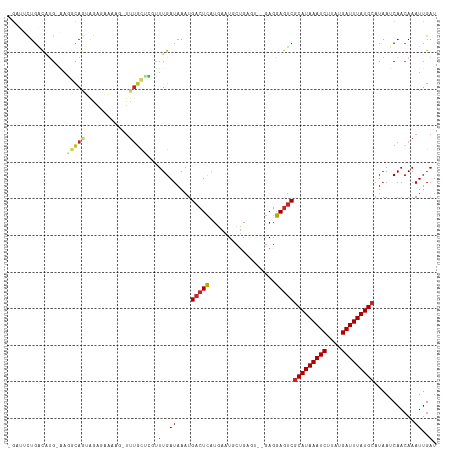
>dm3.chr3L 4958839 112 - 24543557 -GAUACUGACAUG-AAGGCAGUAGAGAAAAG-UUUGCUCGUUUGAUAAAUGACUCAUGAAUGCUGAGU--GAGGAGUCGCAUAAAUCUUAUGAUUUAUGCAUAAUCAACAAAUUGAU -..(((((.(...-...))))))......((-((((.....(((((....(((((....((.....))--...)))))(((((((((....)))))))))...)))))))))))... ( -26.00, z-score = -1.29, R) >droSim1.chr3L 4497106 114 - 22553184 -GAUUCUGACAUG-AAGGCAGUAGAGAAAAG-UUUGCUCGUUUGAUAAAUGACUCAUGAAUGCUGAGUGAGAGGAGUCGCAUAAAUCUUAUGAUUUAUGCAUAAUCAACAAAUUGAU -((((((..(((.-..((((...(((....(-((((.((....)))))))..))).....))))..)))...))))))(((((((((....)))))))))...(((((....))))) ( -29.40, z-score = -1.92, R) >droSec1.super_2 4939294 114 - 7591821 -GAUUCUGACAUG-GAGGCAGUAGAGAAAAG-UUUGCUCGUUUGAUAAAUGACUCAUGAAUGCUGAGUGAGAGGAGUCGCAUAAAUCUUAUGAUUUAUGCAUAAUCAACAAAUUGAU -((((((..(((.-..((((...(((....(-((((.((....)))))))..))).....))))..)))...))))))(((((((((....)))))))))...(((((....))))) ( -29.40, z-score = -1.82, R) >droYak2.chr3L 5541380 112 - 24197627 -GAUUCUGACAUG-AAGGCAGUGGAGAAAAG-UUUGCUCGUUUGAUAAAUGACUCAUAAAUGCUGAGU--GAGGAGUCGCAUAAAUCUUAUGAUUUAUGCAUAAUCAACAAAUUGAU -((((((..(((.-..((((...(((....(-((((.((....)))))))..))).....))))..))--).))))))(((((((((....)))))))))...(((((....))))) ( -30.60, z-score = -2.63, R) >droEre2.scaffold_4784 7670933 112 - 25762168 -GAUUCUGACAUG-AAGGCAGUAGAGAAAAG-UUUGCUCGCUUGAUAAAUGACUCAUAAAUGCUGAGU--GAGGAGUCGCAUAAAUCUUAUGAUUUAUGCAUAAUCAACAAAUUGAU -((((((..(((.-.((((((((((......-)))))).)))).....)))(((((.......)))))--..))))))(((((((((....)))))))))...(((((....))))) ( -31.30, z-score = -2.83, R) >droAna3.scaffold_13337 3050139 110 - 23293914 -GAUUUUGACACG-AAGGCA--AAAAAGGAG-CUGGCCCGUUUGAUAAAUGACUCAUGAAUGCCGAGU--GAGGAGUCGCAUAAAUCUUAUGAUUUAUGCAUAAUCAACAAAUUGAU -((((((..(((.-..((((--.....((.(-....)))(((........))).......))))..))--).))))))(((((((((....)))))))))...(((((....))))) ( -29.20, z-score = -2.60, R) >droWil1.scaffold_180698 1068430 111 + 11422946 -GGUUUUGUUGUGUGACACAGAAAAGUUGUGCUCUCU-GUUUUGAUAAAUGACUUUUGAAUACUGU----GAGAAAACGCAUAAAUCAUAUGAUUUAUGCAUAAUCAAGAAAUUGAU -.(((((.(.......(((((((((((..(..((...-.....))...)..)))))).....))))----)).)))))(((((((((....)))))))))...(((((....))))) ( -26.81, z-score = -2.25, R) >droVir3.scaffold_12758 698884 106 - 972128 CGGGUUUGUC--GUGACACAGAAAACUU----UUUGC-GUUUUGAUAAAUGACUUAUGAAUACUGU----GAGAAGUCGCAUAAAUCUUAUGAUUUAUGCAUAAACAACAAAUUGAU ..(((((((.--((..........((((----((..(-((.((.((((.....)))).)).)).).----.)))))).(((((((((....)))))))))....)).)))))))... ( -26.90, z-score = -2.97, R) >droMoj3.scaffold_6680 1767952 103 - 24764193 -----CUGGCUUGUUUUGCA-AAGUUUU----UCUGCAAUGUUGAUAAAUGACUUAUGAAUGCUGU----UAGAAGUCACAUAAAUCACAUGAUUUAUGCAUAAACAACAAAUUGAU -----..........(((((-.......----..)))))(((((.....((((((.(((......)----)).))))))((((((((....))))))))......)))))....... ( -20.20, z-score = -0.40, R) >droGri2.scaffold_15110 23770987 102 + 24565398 -----CUGGCAUGUGACAUAGAAAACUU----UUUGC--UGUUGAUAAAUGACUUAUGAAUACUGU----GAGAAGUCGCAUAAAUCUUAUGAUUUAUGCAUAAACAACAAAUUGAU -----......(((..........((((----((..(--((((.((((.....)))).))))..).----.)))))).(((((((((....)))))))))....))).......... ( -21.90, z-score = -2.32, R) >consensus _GAUUCUGACAUG_AAGGCAGUAGAGAAAAG_UUUGCUCGUUUGAUAAAUGACUCAUGAAUGCUGAGU__GAGGAGUCGCAUAAAUCUUAUGAUUUAUGCAUAAUCAACAAAUUGAU .........................................(..((....(((((..................)))))(((((((((....)))))))))...........))..). (-14.81 = -14.63 + -0.18)
| Location | 4,958,853 – 4,958,964 |
|---|---|
| Length | 111 |
| Sequences | 9 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 77.91 |
| Shannon entropy | 0.45854 |
| G+C content | 0.34675 |
| Mean single sequence MFE | -19.13 |
| Consensus MFE | -14.47 |
| Energy contribution | -13.94 |
| Covariance contribution | -0.53 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.980447 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
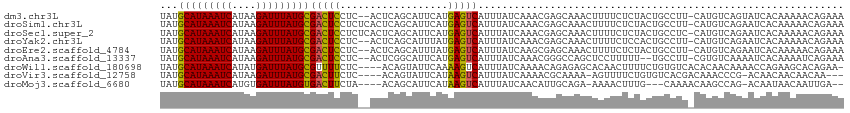
>dm3.chr3L 4958853 111 + 24543557 UAUGCAUAAAUCAUAAGAUUUAUGCGACUCCUC--ACUCAGCAUUCAUGAGUCAUUUAUCAAACGAGCAAACUUUUCUCUACUGCCUU-CAUGUCAGUAUCACAAAAACAGAAA ...(((((((((....)))))))))(((((...--.............))))).....((....(((.((....)))))(((((.(..-...).)))))...........)).. ( -20.09, z-score = -2.27, R) >droSim1.chr3L 4497120 113 + 22553184 UAUGCAUAAAUCAUAAGAUUUAUGCGACUCCUCUCACUCAGCAUUCAUGAGUCAUUUAUCAAACGAGCAAACUUUUCUCUACUGCCUU-CAUGUCAGAAUCACAAAAACAGAAA ...(((((((((....)))))))))(((.......(((((.......)))))............(((((.............)))..)-)..)))................... ( -18.22, z-score = -1.32, R) >droSec1.super_2 4939308 113 + 7591821 UAUGCAUAAAUCAUAAGAUUUAUGCGACUCCUCUCACUCAGCAUUCAUGAGUCAUUUAUCAAACGAGCAAACUUUUCUCUACUGCCUC-CAUGUCAGAAUCACAAAAACAGAAA ...(((((((((....)))))))))(((.......(((((.......)))))............(((((.............)).)))-...)))................... ( -18.92, z-score = -1.75, R) >droYak2.chr3L 5541394 111 + 24197627 UAUGCAUAAAUCAUAAGAUUUAUGCGACUCCUC--ACUCAGCAUUUAUGAGUCAUUUAUCAAACGAGCAAACUUUUCUCCACUGCCUU-CAUGUCAGAAUCACAAAAACAGAAA ...(((((((((....)))))))))(((.....--(((((.......)))))............(((((.............)))..)-)..)))................... ( -18.22, z-score = -1.64, R) >droEre2.scaffold_4784 7670947 111 + 25762168 UAUGCAUAAAUCAUAAGAUUUAUGCGACUCCUC--ACUCAGCAUUUAUGAGUCAUUUAUCAAGCGAGCAAACUUUUCUCUACUGCCUU-CAUGUCAGAAUCACAAAAACAGAAA ...(((((((((....)))))))))(((.....--(((((.......)))))............(((((.............)))..)-)..)))................... ( -18.22, z-score = -0.90, R) >droAna3.scaffold_13337 3050153 109 + 23293914 UAUGCAUAAAUCAUAAGAUUUAUGCGACUCCUC--ACUCGGCAUUCAUGAGUCAUUUAUCAAACGGGCCAGCUCCUUUUU--UGCCUU-CGUGUCAAAAUCACAAAAUCAGAAA ..((((((((((....)))))))))).......--..((((((....(((........)))...(((....)))......--))))..-.(((.......))).......)).. ( -21.20, z-score = -1.54, R) >droWil1.scaffold_180698 1068444 109 - 11422946 UAUGCAUAAAUCAUAUGAUUUAUGCGUUUUCUC----ACAGUAUUCAAAAGUCAUUUAUCAAAACAGAGAGCACAACUUUUCUGUGUCACACAACAAAACCAGAAGCACAGAA- ...(((((((((....)))))))))(((((((.----............................))))))).......((((((((..(............)..))))))))- ( -22.15, z-score = -2.87, R) >droVir3.scaffold_12758 698898 105 + 972128 UAUGCAUAAAUCAUAAGAUUUAUGCGACUUCUC----ACAGUAUUCAUAAGUCAUUUAUCAAAACGCAAAA-AGUUUUCUGUGUCACGACAAACCCG-ACAACAACAACAA--- ...(((((((((....)))))))))(((((...----...........)))))..................-.(((.....((((...........)-))).....)))..--- ( -17.34, z-score = -2.33, R) >droMoj3.scaffold_6680 1767966 103 + 24764193 UAUGCAUAAAUCAUGUGAUUUAUGUGACUUCUA----ACAGCAUUCAUAAGUCAUUUAUCAACAUUGCAGA-AAAACUUUG---CAAAACAAGCCAG-ACAAUAACAAUUGA-- ...(((((((((....)))))))))(((((...----...........)))))...........(((((((-.....))))---)))..........-.((((....)))).-- ( -17.84, z-score = -0.94, R) >consensus UAUGCAUAAAUCAUAAGAUUUAUGCGACUCCUC__ACUCAGCAUUCAUGAGUCAUUUAUCAAACGAGCAAACUUUUCUCUACUGCCUU_CAUGUCAGAAUCACAAAAACAGAAA ...(((((((((....)))))))))(((((..................)))))............................................................. (-14.47 = -13.94 + -0.53)
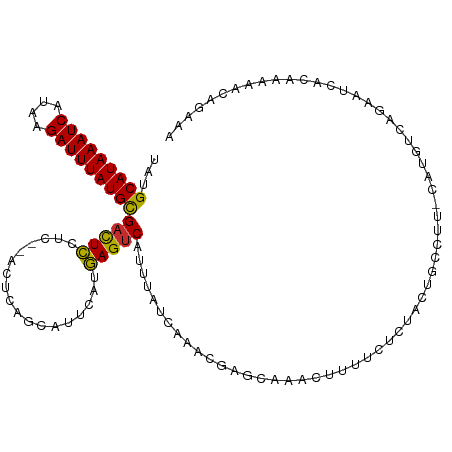
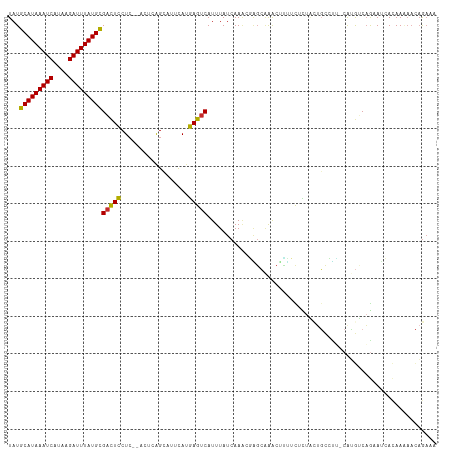
| Location | 4,958,853 – 4,958,964 |
|---|---|
| Length | 111 |
| Sequences | 9 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 77.91 |
| Shannon entropy | 0.45854 |
| G+C content | 0.34675 |
| Mean single sequence MFE | -27.33 |
| Consensus MFE | -14.56 |
| Energy contribution | -14.45 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.643778 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 4958853 111 - 24543557 UUUCUGUUUUUGUGAUACUGACAUG-AAGGCAGUAGAGAAAAGUUUGCUCGUUUGAUAAAUGACUCAUGAAUGCUGAGU--GAGGAGUCGCAUAAAUCUUAUGAUUUAUGCAUA ...(((((((..((.......))..-)))))))..(((.........)))....(((...(.(((((.......)))))--.)...)))(((((((((....)))))))))... ( -28.90, z-score = -1.76, R) >droSim1.chr3L 4497120 113 - 22553184 UUUCUGUUUUUGUGAUUCUGACAUG-AAGGCAGUAGAGAAAAGUUUGCUCGUUUGAUAAAUGACUCAUGAAUGCUGAGUGAGAGGAGUCGCAUAAAUCUUAUGAUUUAUGCAUA .............((((((..(((.-..((((...(((....(((((.((....)))))))..))).....))))..)))...))))))(((((((((....)))))))))... ( -28.60, z-score = -1.18, R) >droSec1.super_2 4939308 113 - 7591821 UUUCUGUUUUUGUGAUUCUGACAUG-GAGGCAGUAGAGAAAAGUUUGCUCGUUUGAUAAAUGACUCAUGAAUGCUGAGUGAGAGGAGUCGCAUAAAUCUUAUGAUUUAUGCAUA ((((((((((..((.......))..-))))))).)))((....((..(((((.(((........))).....)).)))..)).....))(((((((((....)))))))))... ( -28.80, z-score = -1.13, R) >droYak2.chr3L 5541394 111 - 24197627 UUUCUGUUUUUGUGAUUCUGACAUG-AAGGCAGUGGAGAAAAGUUUGCUCGUUUGAUAAAUGACUCAUAAAUGCUGAGU--GAGGAGUCGCAUAAAUCUUAUGAUUUAUGCAUA .............((((((..(((.-..((((...(((....(((((.((....)))))))..))).....))))..))--).))))))(((((((((....)))))))))... ( -29.80, z-score = -2.02, R) >droEre2.scaffold_4784 7670947 111 - 25762168 UUUCUGUUUUUGUGAUUCUGACAUG-AAGGCAGUAGAGAAAAGUUUGCUCGCUUGAUAAAUGACUCAUAAAUGCUGAGU--GAGGAGUCGCAUAAAUCUUAUGAUUUAUGCAUA .............((((((..(((.-.((((((((((......)))))).)))).....)))(((((.......)))))--..))))))(((((((((....)))))))))... ( -30.50, z-score = -2.05, R) >droAna3.scaffold_13337 3050153 109 - 23293914 UUUCUGAUUUUGUGAUUUUGACACG-AAGGCA--AAAAAGGAGCUGGCCCGUUUGAUAAAUGACUCAUGAAUGCCGAGU--GAGGAGUCGCAUAAAUCUUAUGAUUUAUGCAUA .............((((((..(((.-..((((--.....((.(....)))(((........))).......))))..))--).))))))(((((((((....)))))))))... ( -28.40, z-score = -1.85, R) >droWil1.scaffold_180698 1068444 109 + 11422946 -UUCUGUGCUUCUGGUUUUGUUGUGUGACACAGAAAAGUUGUGCUCUCUGUUUUGAUAAAUGACUUUUGAAUACUGU----GAGAAAACGCAUAAAUCAUAUGAUUUAUGCAUA -((((((((.................).)))))))..(((...(((.(.((((..(..........)..)).)).).----)))..)))(((((((((....)))))))))... ( -24.33, z-score = -1.42, R) >droVir3.scaffold_12758 698898 105 - 972128 ---UUGUUGUUGUUGU-CGGGUUUGUCGUGACACAGAAAACU-UUUUGCGUUUUGAUAAAUGACUUAUGAAUACUGU----GAGAAGUCGCAUAAAUCUUAUGAUUUAUGCAUA ---......((((.((-((.(.....).))))))))...(((-(((..(((.((.((((.....)))).)).)).).----.)))))).(((((((((....)))))))))... ( -27.90, z-score = -2.55, R) >droMoj3.scaffold_6680 1767966 103 - 24764193 --UCAAUUGUUAUUGU-CUGGCUUGUUUUG---CAAAGUUUU-UCUGCAAUGUUGAUAAAUGACUUAUGAAUGCUGU----UAGAAGUCACAUAAAUCACAUGAUUUAUGCAUA --(((((....(((((-..(((((......---..)))))..-...))))))))))....((((((.(((......)----)).))))))((((((((....)))))))).... ( -18.70, z-score = 0.48, R) >consensus UUUCUGUUUUUGUGAUUCUGACAUG_AAGGCAGUAGAGAAAAGUUUGCUCGUUUGAUAAAUGACUCAUGAAUGCUGAGU__GAGGAGUCGCAUAAAUCUUAUGAUUUAUGCAUA .............................................................(((((..................)))))(((((((((....)))))))))... (-14.56 = -14.45 + -0.11)

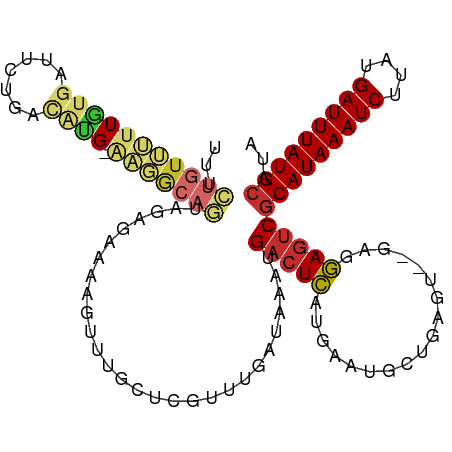
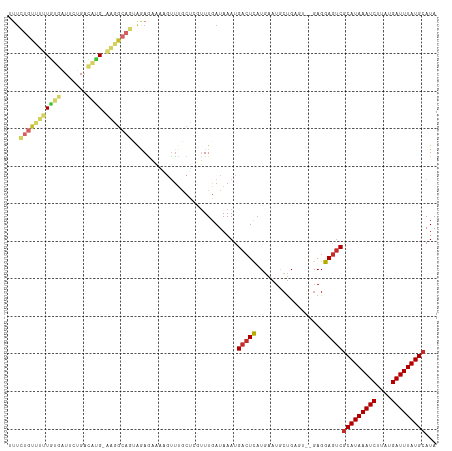
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:03:57 2011