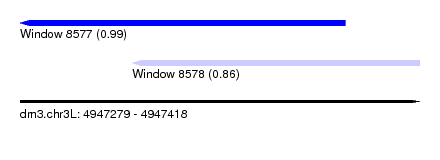
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 4,947,279 – 4,947,418 |
| Length | 139 |
| Max. P | 0.994261 |

| Location | 4,947,279 – 4,947,392 |
|---|---|
| Length | 113 |
| Sequences | 7 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 71.04 |
| Shannon entropy | 0.54217 |
| G+C content | 0.42092 |
| Mean single sequence MFE | -33.68 |
| Consensus MFE | -14.50 |
| Energy contribution | -15.43 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.45 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.68 |
| SVM RNA-class probability | 0.994261 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
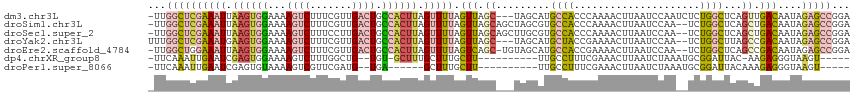
>dm3.chr3L 4947279 113 - 24543557 -UUGGCUCGAAAUUAAGUGGAAAAGUCUUUCGUUGACUGCCACUUAGUUUUAGUUAGC---UAGCAUGCCACCCAAAACUUAAUCCAAUCUCUGGCUCAGUUGACAAUAGAGCCGGA -(((((((((((((((((((...((((.......)))).)))))))))))).((((((---(.(...((((.....................)))).))))))))....))))))). ( -39.40, z-score = -4.43, R) >droSim1.chr3L 4485578 114 - 22553184 -UUGGCUCGAAAUUAAGUGGAAAAGUCUUUCGUUGACUGCCACUUAGUUUUAGUUAGCAGCUAGCGUGCCACCCAAAACUUAAUCCAA--UCUGGCUCAGCUGACAAUAGAGCCGGA -(((((((((((((((((((...((((.......)))).)))))))))))).((((((((((((..((.................)).--.))))))..))))))....))))))). ( -43.13, z-score = -4.72, R) >droSec1.super_2 4927896 114 - 7591821 -UUGGCUCGAAAUUAAGUGGAAAAGUCUUUCCUUGACUGCCACUUAGUUUUAGUUAGCAGCUUGCGUGCCACCCAAAACUUAAUCCAA--UCUGGCUCAGCUGACAAUAGAGCCGGA -(((((((((((((((((((...((((.......)))).)))))))))))).((((((.(....)(.((((.................--..)))).).))))))....))))))). ( -41.81, z-score = -4.46, R) >droYak2.chr3L 5529524 112 - 24197627 UUUGGCUCGAAAUGAAGUGGAAAAGUCUUUCGUUGACUGCCACUUAGUUUUAGUUAGC---UAGCAUGCUACCGAAAACUUAAUCCAA--UCUGGCUUAGCCGACAAUAGAGCCGGA (((((((((((((.((((((...((((.......)))).)))))).))))).((..((---((((........((........))...--....)).))))..))....)))))))) ( -34.96, z-score = -2.77, R) >droEre2.scaffold_4784 7659919 113 - 25762168 -UUGGCUGGAAAUUAAGUGGAAAAGUCUUUCGUUGACUGCCACUUAGUUUUAGUCAGC-UGUAGCAUGCCACCGAAAACUUAAUCCAA--UCUGGCUCAGCCGACAAUAGAGCCGGA -((((((.((((((((((((...((((.......)))).)))))))))))).(((.((-((.(((........((........))...--....))))))).))).....)))))). ( -38.76, z-score = -3.36, R) >dp4.chrXR_group8 3104385 97 - 9212921 -UUCAAAUUGAAUCGAGUGGAAAAGUCUUUGGCUG--UGU-GCUUUGCUUUGCUU----------UUGCCUUUCGAAACUUAAUCUAAAUGCGGAUUAC-AAGAGGGUAAGU----- -.((...(((..(((((.((((((((....(((..--...-.....)))..))))----------)).)).)))))....((((((......)))))))-))...)).....----- ( -18.00, z-score = 0.79, R) >droPer1.super_8066 74 93 + 1396 -UUCAAAUUGAAUCGAGUGUAAAAGUCGUUCGAUG--UGA------GCUUUGCUU----------UUGCCUUUCGAAACUUAAUCUAAAUGCGGAUUACAAAGAGGGUAAGU----- -((((.((((((.(((.(.....).))))))))).--)))------).......(----------((((((((.......((((((......))))))....))))))))).----- ( -19.70, z-score = -0.64, R) >consensus _UUGGCUCGAAAUUAAGUGGAAAAGUCUUUCGUUGACUGCCACUUAGUUUUAGUUAGC___UAGCAUGCCACCCAAAACUUAAUCCAA__UCUGGCUCAGCUGACAAUAGAGCCGGA ...((((((((((.((((((...((((.......)))).)))))).))))).(((.((.........((((.....................))))...)).)))....)))))... (-14.50 = -15.43 + 0.92)
| Location | 4,947,318 – 4,947,418 |
|---|---|
| Length | 100 |
| Sequences | 7 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 68.56 |
| Shannon entropy | 0.58177 |
| G+C content | 0.40004 |
| Mean single sequence MFE | -28.20 |
| Consensus MFE | -8.95 |
| Energy contribution | -9.71 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.859932 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
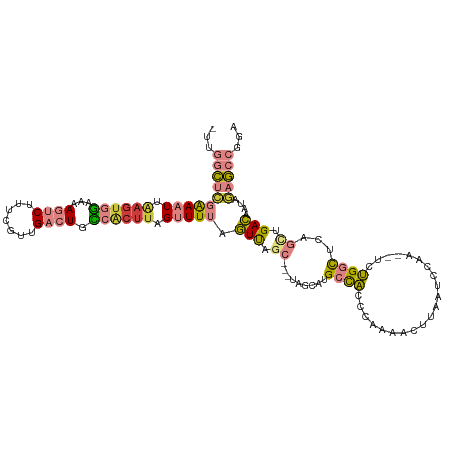
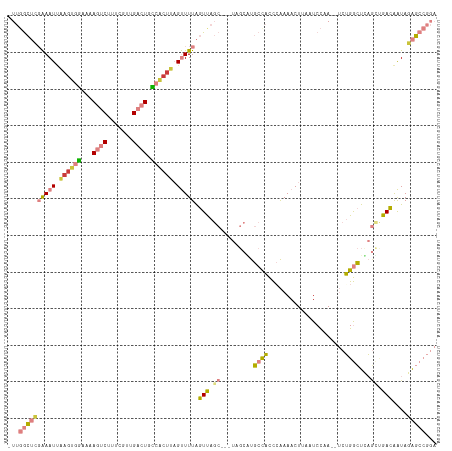
>dm3.chr3L 4947318 100 - 24543557 UUAAACGAAGAGCGAAGGAAAA---UCGCUUGGCUCGAAAUUAAGUGGAAAAGUCUUUCGUUGACUGC--CACUUAGUUUUAGUU---AGCUAGCAUGCCACCCAAAA ...(((...((((.((((....---.).))).))))((((((((((((...((((.......)))).)--))))))))))).)))---.((......))......... ( -30.80, z-score = -3.02, R) >droSim1.chr3L 4485615 103 - 22553184 UUAAACGAAGAGCGAAGGAAAA---UCGCUUGGCUCGAAAUUAAGUGGAAAAGUCUUUCGUUGACUGC--CACUUAGUUUUAGUUAGCAGCUAGCGUGCCACCCAAAA .........((((((.......---))))))(((.(((((((((((((...((((.......)))).)--))))))))))).(((((...)))))).)))........ ( -34.40, z-score = -3.37, R) >droSec1.super_2 4927933 103 - 7591821 UUAAACGAAGAGCGAAGGAAAA---UCGCUUGGCUCGAAAUUAAGUGGAAAAGUCUUUCCUUGACUGC--CACUUAGUUUUAGUUAGCAGCUUGCGUGCCACCCAAAA .........((((((.......---))))))(((.(((((((((((((...((((.......)))).)--))))))))))).....((.....))).)))........ ( -33.20, z-score = -3.06, R) >droYak2.chr3L 5529561 103 - 24197627 UUAAAUGAAAAGCAAAGGAAAAUCGUCGUUUGGCUCGAAAUGAAGUGGAAAAGUCUUUCGUUGACUGC--CACUUAGUUUUAGUU---AGCUAGCAUGCUACCGAAAA ............((((.((......)).))))(((.(((((.((((((...((((.......)))).)--))))).))))))))(---(((......))))....... ( -24.10, z-score = -1.07, R) >droEre2.scaffold_4784 7659956 102 - 25762168 UUAAACGAAGAGCGAAGGAAAA---GCGCUUGGCUGGAAAUUAAGUGGAAAAGUCUUUCGUUGACUGC--CACUUAGUUUUAGUCAGCUG-UAGCAUGCCACCGAAAA .....((....((.........---))(((((((((((((((((((((...((((.......)))).)--)))))))))))...))))))-.))).......)).... ( -31.20, z-score = -2.13, R) >dp4.chrXR_group8 3104418 92 - 9212921 UUAAACCAAAUUGAAGAGGGAAAAGUACUUCAAAUUG-AAUCGAGUGGAAAAGUCUUUGGCUGUGUGCUUUGCUUUGCUUUUGCCUUUCGAAA--------------- ......(((.((((((...........)))))).)))-..(((((.((((((((....(((..........)))..)))))).)).)))))..--------------- ( -19.40, z-score = -0.25, R) >droPer1.super_8066 108 87 + 1396 UUAAACCAAAUUGAAGAGGGAAAAGUACUUCAAAUUG-AAUCGAGUGUAAAAGUCGUUCGAUGUG-----AGCUUUGCUUUUGCCUUUCGAAA--------------- ..........((((.((((.(((((((.((((.((((-((.(((.(.....).))))))))).))-----))...))))))).))))))))..--------------- ( -24.30, z-score = -3.06, R) >consensus UUAAACGAAGAGCGAAGGAAAA___UCGCUUGGCUCGAAAUUAAGUGGAAAAGUCUUUCGUUGACUGC__CACUUAGUUUUAGUUAGCAGCUAGCAUGCCACCCAAAA .........((((.(((...........))).))))(((((.(((((....((((.......))))....))))).)))))........................... ( -8.95 = -9.71 + 0.76)

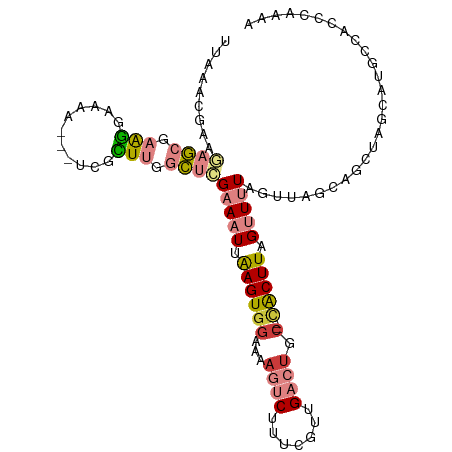
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:03:51 2011