| Sequence ID | dm3.chr3L |
|---|---|
| Location | 4,920,810 – 4,920,911 |
| Length | 101 |
| Max. P | 0.781524 |

| Location | 4,920,810 – 4,920,911 |
|---|---|
| Length | 101 |
| Sequences | 9 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 67.14 |
| Shannon entropy | 0.64414 |
| G+C content | 0.51714 |
| Mean single sequence MFE | -22.53 |
| Consensus MFE | -13.74 |
| Energy contribution | -14.30 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.47 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.781524 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 4920810 101 - 24543557 AGAGUGGAACGGUGCACCGGGGAAUACUUUGCCAAAUUGCACAAUCUACUCAGUCU-ACUCAGCCCAACCC-----------CUGACUUCAGUCCGAAUCCUUUUUCCCCAGG .(((((((...(((((...((.((....)).))....)))))..))))))).....-.............(-----------(((((....))).(((......)))...))) ( -23.60, z-score = -0.32, R) >droSim1.chr3L 4459039 101 - 22553184 AGAGUGGAACGGUGCACCGGGGAAUACUUUGCCAAAUUGCACAAUCUACUCAGUCU-ACUCAGCCCAACCC-----------CUGACUCCAGUCCGAAUCCUUUUUCCCCAGG .(((((((...(((((...((.((....)).))....)))))..))))))).....-.............(-----------(((((....))).(((......)))...))) ( -23.60, z-score = -0.29, R) >droSec1.super_2 4901317 100 - 7591821 AGAGUGGAACGGUGCACC-GGGAAUACUUUGCCAAAUUGCACAAUCUACUCAGUCU-ACUCAGCCCAACCC-----------CUGACUCCAGUCCGAAUCCUUUUUCCCCAGG .(((((((...(((((..-((.((....)).))....)))))..))))))).....-.............(-----------(((((....))).(((......)))...))) ( -22.20, z-score = -0.42, R) >droYak2.chr3L 5501048 113 - 24197627 AGAGUAGAACGGUGCACCGGGGAAUACUUUGCCAAAUUGCACAAUCUACUCAGUCUCAUGUCCUCUGAACUGCGCCCAGUCUCCGACUCCAGUACGAAUCCUUCUUCCCCAGG .(((((((...(((((...((.((....)).))....)))))..))))))).((((...(((....((.(((....)))))...)))...)).)).................. ( -26.20, z-score = -0.09, R) >droEre2.scaffold_4784 7632964 100 - 25762168 AGAGUGGAACGGUGCACCGGGGAAUACUUUGCCAAAUUGCACAAUCUACUCAGUCUCAUGUCCCCCGAACU-------------GACUCCAGUCCGAAUCCUUCUUCCCCAGG .(((((((...(((((...((.((....)).))....)))))..)))))))..............((.(((-------------(....)))).))................. ( -25.40, z-score = -0.43, R) >droAna3.scaffold_13337 3010222 105 - 23293914 AGAGUAGAACGGUGCACCGGGGAAUACUUUGCCAAAUUGCACAAUCUACUCUGUCUCCUGUCCGAACUCCC--------CUUCUCCUGCCAGAGGACUUUCCCCAUGUCCAGA ((((((((...(((((...((.((....)).))....)))))..)))))))).....(((..((((.....--------.)))((((.....))))..........)..))). ( -27.40, z-score = -0.80, R) >droPer1.super_440 290 79 - 23054 AGAGUCGAACGGUGCACCGGGGAAUACUUUGCCAAAUUGCACAAUCUACUCGGUCUUCGGCCCCCAAUCCU--------CUUCUAUU-------------------------- .((((.((...(((((...((.((....)).))....)))))..)).))))((.(....))).........--------........-------------------------- ( -19.40, z-score = -0.82, R) >dp4.chrXR_group8 3073520 79 - 9212921 AGAGUCGAACGGUGCACCGGGGAAUACUUUGCCAAAUUGCACAAUCUACUCGGUCUUCGGGCCCCAAUCCU--------CUUCUAUU-------------------------- .((((.((...(((((...((.((....)).))....)))))..)).))))((((....))))........--------........-------------------------- ( -23.80, z-score = -2.00, R) >droVir3.scaffold_12758 639128 83 - 972128 AUAGCCAAACGGUGCACCGGGGAAUACUUUACCAAAUUGCACUACCCAGUCUCACAGUCAACUCCUGCUCC-------AACUCAAGCUCC----------------------- ..(((.....((((((...((..........))....))))))...........(((.......)))....-------.......)))..----------------------- ( -11.20, z-score = 0.97, R) >consensus AGAGUGGAACGGUGCACCGGGGAAUACUUUGCCAAAUUGCACAAUCUACUCAGUCU_ACGCCCCCCAACCC__________UCUGACUCCAGUCCGAAUCCUU_UUCCCCAGG .((((.((...(((((...((.((....)).))....)))))..)).)))).............................................................. (-13.74 = -14.30 + 0.56)

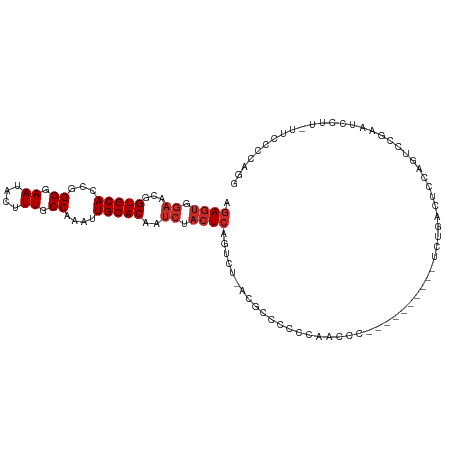

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:03:49 2011