
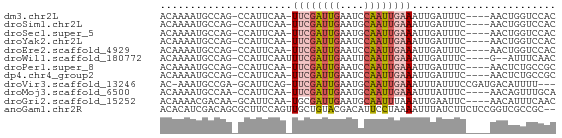
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 4,326,289 – 4,326,349 |
| Length | 60 |
| Max. P | 0.693961 |

| Location | 4,326,289 – 4,326,349 |
|---|---|
| Length | 60 |
| Sequences | 12 |
| Columns | 66 |
| Reading direction | reverse |
| Mean pairwise identity | 77.71 |
| Shannon entropy | 0.48662 |
| G+C content | 0.36153 |
| Mean single sequence MFE | -12.70 |
| Consensus MFE | -4.66 |
| Energy contribution | -4.94 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.693961 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
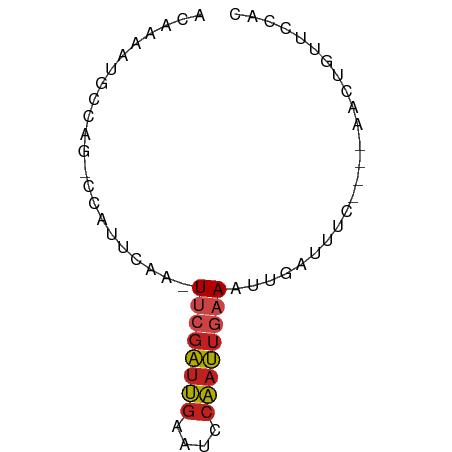
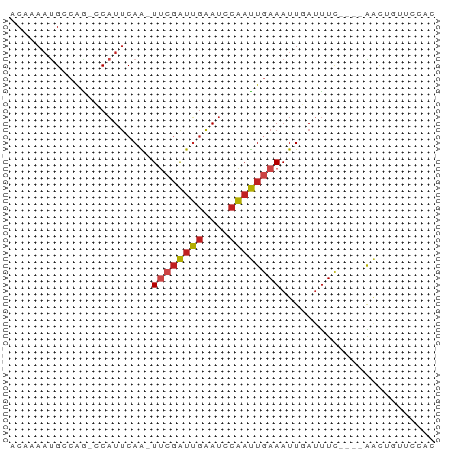
>dm3.chr2L 4326289 60 - 23011544 ACAAAAUGCCAG-CCAUUCAA-UUCGAUUGAAUCCAAUUGAAAUUGAUUUC----AACUGGUCCAC .......(((((-....((((-((((((((....)))))).))))))....----..))))).... ( -15.00, z-score = -3.09, R) >droSim1.chr2L 4248465 60 - 22036055 ACAAAAUGCCAG-CCAUUCAA-UUCGAUUGAAUGCAAUUGAAAUUGAUUUC----AACUGGUCCAC .......(((((-....((((-((((((((....)))))).))))))....----..))))).... ( -14.80, z-score = -2.51, R) >droSec1.super_5 2423852 60 - 5866729 ACAAAAUGCCAG-CCAUUCAA-UUCGAUUGAAUGCAAUUGAAAUUGAUUUC----AACUGGUCCAC .......(((((-....((((-((((((((....)))))).))))))....----..))))).... ( -14.80, z-score = -2.51, R) >droYak2.chr2L 4357770 60 - 22324452 ACAAAAUGCCAG-CCAUUCAA-UUCGAUUGAAUCCAAUUGAAAUUGAUUUC----AACUGGUCCAC .......(((((-....((((-((((((((....)))))).))))))....----..))))).... ( -15.00, z-score = -3.09, R) >droEre2.scaffold_4929 4403211 60 - 26641161 ACAAAAUGCCAG-CCAUUCAA-UUCGAUUGAAUCCAAUUGAAAUUGAUUUC----AACUGGUCCAC .......(((((-....((((-((((((((....)))))).))))))....----..))))).... ( -15.00, z-score = -3.09, R) >droWil1.scaffold_180772 2051134 59 - 8906247 ACAAAAUGCCAG-CCAUUCAAUUUCGAUUGAAUUCAAUUGAAAUUGAUUUC----G--AUUUCAAC ............-....(((((((((((((....)))))))))))))....----.--........ ( -14.30, z-score = -3.15, R) >droPer1.super_8 398945 60 + 3966273 ACAAAAUGCCAG-CCAUUCAA-UUCGAUUGAAUCCAAUUGAAAUUGAUUUC----AACUCUGCCGC .......(.(((-....((((-((((((((....)))))).))))))....----....))).).. ( -9.50, z-score = -1.21, R) >dp4.chr4_group2 322332 60 - 1235136 ACAAAAUGCCAG-CCAUUCAA-UUCGAUUGAAUCCAAUUGAAAUUGAUUUC----AACUCUGCCGC .......(.(((-....((((-((((((((....)))))).))))))....----....))).).. ( -9.50, z-score = -1.21, R) >droVir3.scaffold_13246 394901 60 - 2672878 AC-AAAUGCCGA-GCAUUCAG-UUCGAUUGAAUGCAAUUGAAAUUUAUUUCCGAUGACAUUUU--- ..-(((((.((.-((((((((-(...)))))))))....((((....))))...)).))))).--- ( -11.30, z-score = -1.24, R) >droMoj3.scaffold_6500 396553 60 - 32352404 ACAAAAUGCCAA-CCAUUCAA-UUCGAUUGAAUGCAAUUGAAAUUUAUUUC----AACAGUUUGCA ......(((.((-((((((((-(...))))))))...((((((....))))----))..))).))) ( -11.00, z-score = -1.75, R) >droGri2.scaffold_15252 346334 60 + 17193109 ACAAAACGACAA-GCAUUCAA-UGCGAUUGAAUGCAAUUUAAAUUGAAUUC----AACAUUUCAAC ............-((((((((-(...)))))))))........(((((...----.....))))). ( -11.80, z-score = -2.71, R) >anoGam1.chr2R 22987190 64 - 62725911 ACACAUCGACAGCGCUUCCAGUUGCUGUACGACAUUCCUAAAAUUUAUCUUCUCCGGUCGCCGC-- .....((((((((((.....).)))))).)))......................(((...))).-- ( -10.40, z-score = -0.45, R) >consensus ACAAAAUGCCAG_CCAUUCAA_UUCGAUUGAAUCCAAUUGAAAUUGAUUUC____AACUGUUCCAC ......................((((((((....))))))))........................ ( -4.66 = -4.94 + 0.28)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:16:10 2011