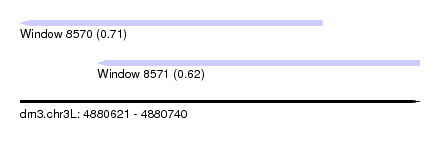
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 4,880,621 – 4,880,740 |
| Length | 119 |
| Max. P | 0.706654 |

| Location | 4,880,621 – 4,880,711 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 77.70 |
| Shannon entropy | 0.43635 |
| G+C content | 0.37913 |
| Mean single sequence MFE | -19.57 |
| Consensus MFE | -12.88 |
| Energy contribution | -12.89 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.706654 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
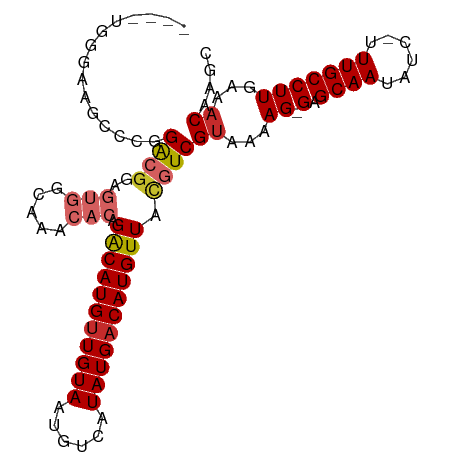
>dm3.chr3L 4880621 90 - 24543557 ------GACAUGUUGUAAUGUCAUAUGACAUGUUACGUCGUAAAAG-GA--GCAAUAUC-UUUGCCUUGAAACAAGCGAGCAAAAGGAU--GCACGAGAAAC----------- ------(((((((..((......))..)))))))...((((...((-(.--((((....-.)))))))...........(((......)--)))))).....----------- ( -20.20, z-score = -1.21, R) >droSim1.chr3L 4419770 90 - 22553184 ------GACAUGUUGUAAUGUCAUAUGACAUGUUACGUCGUAAAAG-GA--GCAAUAUC-UUUGCCUUGAAACAAGCGAGCAAAAGGAU--GCACGAGAAAC----------- ------(((((((..((......))..)))))))...((((...((-(.--((((....-.)))))))...........(((......)--)))))).....----------- ( -20.20, z-score = -1.21, R) >droSec1.super_2 4867678 90 - 7591821 ------GACAUGUUGUAAUGUCAUAUGACAUGUUACGUCGUAAAAG-GA--GCAAUAUC-UUUGCCUUGAAACAAGCGAGCAAAAGGAU--GCACGAGAAAC----------- ------(((((((..((......))..)))))))...((((...((-(.--((((....-.)))))))...........(((......)--)))))).....----------- ( -20.20, z-score = -1.21, R) >droYak2.chr3L 5465825 90 - 24197627 ------GACAUGUUGUAAUGUCAUAUGACAUGUUACGUCGUAAAAG-GA--GCAAUAUC-UUUGCCUUGAAACAAGCGAGCAAAAGGAU--GCACGAAAAAU----------- ------(((((((..((......))..)))))))...((((...((-(.--((((....-.)))))))...........(((......)--)))))).....----------- ( -20.20, z-score = -1.39, R) >droEre2.scaffold_4784 7599153 90 - 25762168 ------GACAUGUUGUAAUGUCAUAUGACAUGUUACGUCGUAAAAG-GA--GCAAUAUC-UUUGCCUUGAAACAAGCGAGCAGAAGGAU--GCACGAAAAAU----------- ------(((((((..((......))..)))))))...((((...((-(.--((((....-.)))))))...........(((......)--)))))).....----------- ( -21.00, z-score = -1.46, R) >droAna3.scaffold_13337 2973851 93 - 23293914 ------GGCAUGCUGUAAUGUCAUAUGACAUGUUACGUCGUAAAAGGA---GCAAUAUC-UUUGCCUUGAAACAAGCAGAAAGAAGG-----AAAAGAAGUAAAAGAU----- ------(((((......))))).((((((.......))))))......---......((-(((.((((...............))))-----.)))))..........----- ( -15.66, z-score = -0.29, R) >dp4.chrXR_group8 3033725 101 - 9212921 ------GACAUGUUGUAAUGUCAUAUGACAUGUUACGUCGUAAAAG-GA--GCAAUAUC-UUUGCCUUGAAACAAGCAAAAAGAAAAGUCCGAACCAGAGCCAGAACCAGA-- ------(((((......))))).((((((.......))))))...(-(.--.(....((-(((..((((...))))...)))))...(..(......)..)..)..))...-- ( -17.00, z-score = -0.47, R) >droPer1.super_24 1538795 101 + 1556852 ------GACAUGUUGUAAUGUCAUAUGACAUGUUACGUCGUAAAAG-GA--GCAAUAUC-UUUGCCUUGAAACAAGCAAAAAGAAAAGUCCGAACCAGAGCCAGAACCAGA-- ------(((((......))))).((((((.......))))))...(-(.--.(....((-(((..((((...))))...)))))...(..(......)..)..)..))...-- ( -17.00, z-score = -0.47, R) >droWil1.scaffold_180698 986037 90 + 11422946 ------GACAUGUUGUAAUGUCAUAUGACAUGUUACGUCGUAAAAGCGA--GCAAUAUCUUUUGCCUU----CUUUCUAAAAGAAAAG---GAAGCAGCAACAGC-------- ------(((((((..((......))..)))))))...((((....))))--.......((.((((...----((((((........))---))))..)))).)).-------- ( -20.40, z-score = -1.38, R) >droVir3.scaffold_12758 594623 97 - 972128 CUCACGGACAUGUUGUAAUGUCAUAUGACAUGUUAUGUCGUAAAAGCGA--GCAAUAUC-UUUGCCUUGAAACAAG---AAAGAAAAC----AAGUGAAAAGAAAGU------ .((((.(((((((..((......))..))))))).((((((....))))--.))...((-(((..((((...))))---)))))....----..)))).........------ ( -20.60, z-score = -1.78, R) >droMoj3.scaffold_6680 1671033 90 - 24764193 ------GACAUGUUGUAAUGUCAUAUGACAUGUUAUGUCGUAAAAGCGA--GCAAUAUC-UUUGCCUUGAAACAAG---AAAGAAAAC-----AGUAGAGAAAAAGU------ ------(((((((..((......))..))))))).((((((....))))--.))...((-(((((((((...))))---...(....)-----.)))))))......------ ( -18.30, z-score = -2.08, R) >droGri2.scaffold_15110 23682369 110 + 24565398 --CACGCACAUAUUGUAAUGUCAUAUGACAUGUUAUGUCGUAAAAGCGAACGCAAUAUC-UUUGCCUUGAAACAAGUGCAAAGAAAACACACAAUCAGAGAUAGAGUUAGAGU --.(((.(((((.....(((((....)))))..))))))))....((....))....((-(((((((((...)))).)))))))............................. ( -24.10, z-score = -1.52, R) >consensus ______GACAUGUUGUAAUGUCAUAUGACAUGUUACGUCGUAAAAG_GA__GCAAUAUC_UUUGCCUUGAAACAAGCGAAAAGAAAAAU__GAACGAGAAACA_A________ ......(((((......))))).((((((.......)))))).........((((......))))................................................ (-12.88 = -12.89 + 0.01)

| Location | 4,880,644 – 4,880,740 |
|---|---|
| Length | 96 |
| Sequences | 9 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 84.70 |
| Shannon entropy | 0.33213 |
| G+C content | 0.43078 |
| Mean single sequence MFE | -25.28 |
| Consensus MFE | -17.19 |
| Energy contribution | -18.02 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.619430 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 4880644 96 - 24543557 ----UGGAAAGCCCGGACGGAGUGGCAAACACAGACAUGUUGUAAUGUCAUAUGACAUGUUACGUCGUAAAAG-GAGCAAUAUC-UUUGCCUUGAAACAAGC ----.((.....)).((((..(((.....))).(((((((..((......))..))))))).))))((...((-(.((((....-.)))))))...)).... ( -26.00, z-score = -1.75, R) >droSim1.chr3L 4419793 96 - 22553184 ----UGGGAAGCCCGGACGGAGUGGCAAACACAGACAUGUUGUAAUGUCAUAUGACAUGUUACGUCGUAAAAG-GAGCAAUAUC-UUUGCCUUGAAACAAGC ----.(((...))).((((..(((.....))).(((((((..((......))..))))))).))))((...((-(.((((....-.)))))))...)).... ( -27.70, z-score = -2.07, R) >droSec1.super_2 4867701 96 - 7591821 ----UGGGAAGCCGGGACGGAGUGGCACACACAGACAUGUUGUAAUGUCAUAUGACAUGUUACGUCGUAAAAG-GAGCAAUAUC-UUUGCCUUGAAACAAGC ----.((....))..((((..(((.....))).(((((((..((......))..))))))).))))((...((-(.((((....-.)))))))...)).... ( -27.40, z-score = -1.80, R) >droYak2.chr3L 5465848 96 - 24197627 ----UGGGAAGUCCGGACGGAGUGGCAAACACAGACAUGUUGUAAUGUCAUAUGACAUGUUACGUCGUAAAAG-GAGCAAUAUC-UUUGCCUUGAAACAAGC ----..((....)).((((..(((.....))).(((((((..((......))..))))))).))))((...((-(.((((....-.)))))))...)).... ( -26.60, z-score = -1.87, R) >droEre2.scaffold_4784 7599176 95 - 25762168 -----UGGAAGCCCGGACGGAGUGGCAAACACAGACAUGUUGUAAUGUCAUAUGACAUGUUACGUCGUAAAAG-GAGCAAUAUC-UUUGCCUUGAAACAAGC -----.((....)).((((..(((.....))).(((((((..((......))..))))))).))))((...((-(.((((....-.)))))))...)).... ( -26.00, z-score = -1.74, R) >droAna3.scaffold_13337 2973877 100 - 23293914 UAGUCUAAAAAUCCGGACGGAGUGGCAUACACAGGCAUGCUGUAAUGUCAUAUGACAUGUUACGUCGUAAAAG-GAGCAAUAUC-UUUGCCUUGAAACAAGC ...........(((..(((..(((((((..((((.....))))...(((....))))))))))..)))....)-))((((....-.))))((((...)))). ( -24.20, z-score = -1.01, R) >dp4.chrXR_group8 3033759 98 - 9212921 --ACACACAGACAUAGACGGAGUGGCAAGCACAGACAUGUUGUAAUGUCAUAUGACAUGUUACGUCGUAAAAG-GAGCAAUAUC-UUUGCCUUGAAACAAGC --.............((((..(((.....))).(((((((..((......))..))))))).))))((...((-(.((((....-.)))))))...)).... ( -24.30, z-score = -1.34, R) >droWil1.scaffold_180698 986060 86 + 11422946 --------------AGAGAGAUACGGACACACAGACAUGUUGUAAUGUCAUAUGACAUGUUACGUCGUAAAAGCGAGCAAUAUCUUUUGCCUUCUUUCUA-- --------------(((((((....(((.....(((((((..((......))..)))))))..)))(((((((..(....)..)))))))..))))))).-- ( -22.90, z-score = -2.19, R) >droMoj3.scaffold_6680 1671055 95 - 24764193 --ACACACAGACGCUGGGGCA----CACGCACAGACAUGUUGUAAUGUCAUAUGACAUGUUAUGUCGUAAAAGCGAGCAAUAUC-UUUGCCUUGAAACAAGA --.........((((......----.(((.((((((((((..((......))..))))))).))))))...)))).((((....-.))))((((...)))). ( -22.40, z-score = -0.13, R) >consensus ____UGGGAAGCCCGGACGGAGUGGCAAACACAGACAUGUUGUAAUGUCAUAUGACAUGUUACGUCGUAAAAG_GAGCAAUAUC_UUUGCCUUGAAACAAGC ...............((((..(((.....))).(((((((((((......))))))))))).))))..........((((......))))............ (-17.19 = -18.02 + 0.83)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:03:45 2011