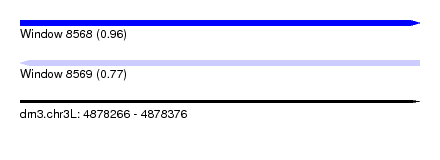
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 4,878,266 – 4,878,376 |
| Length | 110 |
| Max. P | 0.962049 |

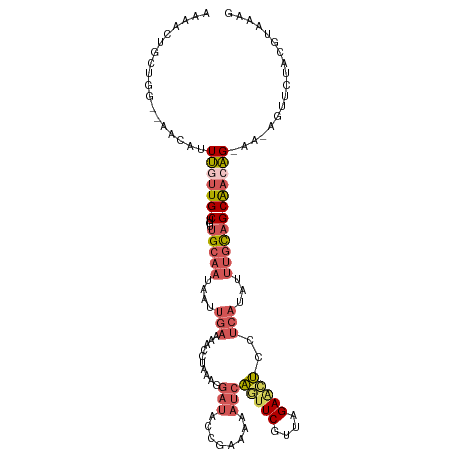
| Location | 4,878,266 – 4,878,376 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 66.11 |
| Shannon entropy | 0.60458 |
| G+C content | 0.36063 |
| Mean single sequence MFE | -24.48 |
| Consensus MFE | -8.48 |
| Energy contribution | -10.16 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.962049 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 4878266 110 + 24543557 AAAACUGCUAGAGAACAUUUGUUGCUAUUGCAAUAAUUGAAAACCCAAAGGAUACAAAAAAAUCAGUUCGUUAGAACUCCUCAUAUUUGCAGCAACAG------UUCUACGUAAAG .....(((...(((((...(((((((...((((....(((...((....)).............(((((....)))))..)))...))))))))))))------))))..)))... ( -23.40, z-score = -2.70, R) >droAna3.scaffold_13337 2971376 105 + 23293914 AUGAUUCCCAA------AUAAUUGCUCAUG---UAAAUAAAAACUUGAAUAAUUCAGUUUGCAACUUUCGUUGGAUGGCAGCCCUCUUGU-GCAUCUG-AAUAGUUUCAAACAAAG ...........------..(((((.(((.(---(........)).))).)))))..(((((.((((....(..((((((((.....))))-.))))..-)..)))).))))).... ( -18.60, z-score = -0.66, R) >droYak2.chr3L 5463399 96 + 24197627 GAAGCUAUAGG--AACCUUUCUUGCUGUUGCAAUAAUUGAAAACAUAAGGGAUACCAACAAAUCAGUUCGUUAGAGCUCCUCAUAUUUGCAGCAAAAG------------------ ..(((...(((--(....)))).)))(((((((....(((........((....))........(((((....)))))..)))...))))))).....------------------ ( -20.60, z-score = -1.33, R) >droSec1.super_2 4865365 113 + 7591821 AAAACUGCUGA---ACAUUUGUUGCCGUUGCAAUAAUUGAAAACCUAAAGGAUACCGAAAAAUCAGUUCGUUAGAACUCCUCAUAUUUGCAGCGACAGCAACAGUUCUACGUAAAG .....(((.((---((..((((((.((((((((....(((...((....)).............(((((....)))))..)))...)))))))).))))))..))))...)))... ( -29.60, z-score = -3.62, R) >droSim1.chr3L 4417416 114 + 22553184 AAAACUGCUGG--AACAUUUGUUGCUGUUGCAAUAAUUGAAACCCUAAAGGAUACCGAAAAAUCAGUUCGUUAGAAAUCCUCAUAUUUGCAGCAACAGCAACAGUUCUACGUAAAG .....((((((--(((...((((((((((((.................(((((...((....))..(((....))))))))((....))..)))))))))))))))))).)))... ( -30.20, z-score = -3.64, R) >consensus AAAACUGCUGG__AACAUUUGUUGCUGUUGCAAUAAUUGAAAACCUAAAGGAUACCGAAAAAUCAGUUCGUUAGAACUCCUCAUAUUUGCAGCAACAG_AA_AGUUCUACGUAAAG ..................(((((((...(((((....(((..........(((........)))(((((....)))))..)))...)))))))))))).................. ( -8.48 = -10.16 + 1.68)

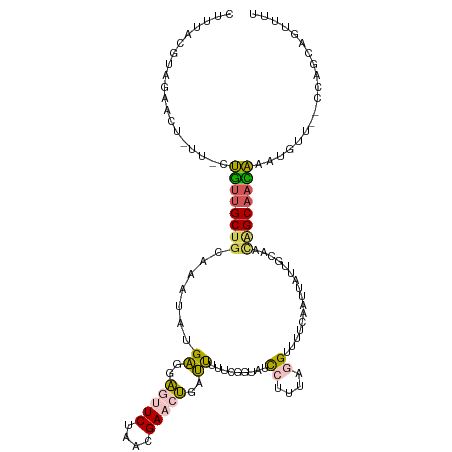
| Location | 4,878,266 – 4,878,376 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 66.11 |
| Shannon entropy | 0.60458 |
| G+C content | 0.36063 |
| Mean single sequence MFE | -25.98 |
| Consensus MFE | -9.92 |
| Energy contribution | -9.96 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.769345 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 4878266 110 - 24543557 CUUUACGUAGAA------CUGUUGCUGCAAAUAUGAGGAGUUCUAACGAACUGAUUUUUUUGUAUCCUUUGGGUUUUCAAUUAUUGCAAUAGCAACAAAUGUUCUCUAGCAGUUUU ........((((------((((((.((((((...((..(((((....)))))..))..))))))......(((....((.((.((((....)))).)).))..))))))))))))) ( -24.70, z-score = -1.39, R) >droAna3.scaffold_13337 2971376 105 - 23293914 CUUUGUUUGAAACUAUU-CAGAUGC-ACAAGAGGGCUGCCAUCCAACGAAAGUUGCAAACUGAAUUAUUCAAGUUUUUAUUUA---CAUGAGCAAUUAU------UUGGGAAUCAU .....((((((...(((-(((.(((-......(((......)))(((....))))))..))))))..))))))......((((---.((((....))))------.))))...... ( -18.10, z-score = 0.34, R) >droYak2.chr3L 5463399 96 - 24197627 ------------------CUUUUGCUGCAAAUAUGAGGAGCUCUAACGAACUGAUUUGUUGGUAUCCCUUAUGUUUUCAAUUAUUGCAACAGCAAGAAAGGUU--CCUAUAGCUUC ------------------(((((..((.((((((((((....((((((((....))))))))....)))))))))).))....((((....)))))))))...--........... ( -22.40, z-score = -1.31, R) >droSec1.super_2 4865365 113 - 7591821 CUUUACGUAGAACUGUUGCUGUCGCUGCAAAUAUGAGGAGUUCUAACGAACUGAUUUUUCGGUAUCCUUUAGGUUUUCAAUUAUUGCAACGGCAACAAAUGU---UCAGCAGUUUU ......((.(((((((((((((.((((.((((..(((((..((....))(((((....))))).)))))...)))).))......)).)))))))))...))---)).))...... ( -31.20, z-score = -2.54, R) >droSim1.chr3L 4417416 114 - 22553184 CUUUACGUAGAACUGUUGCUGUUGCUGCAAAUAUGAGGAUUUCUAACGAACUGAUUUUUCGGUAUCCUUUAGGGUUUCAAUUAUUGCAACAGCAACAAAUGUU--CCAGCAGUUUU ......((.((((((((((((((((((.((((..((((((.((....))(((((....)))))))))))....))))))......))))))))))))...)))--)..))...... ( -33.50, z-score = -3.45, R) >consensus CUUUACGUAGAACU_UU_CUGUUGCUGCAAAUAUGAGGAGUUCUAACGAACUGAUUUUUCGGUAUCCUUUAGGUUUUCAAUUAUUGCAACAGCAACAAAUGUU__CCAGCAGUUUU ...................((((((((.......((..(((((....)))))..)).........((....))................))))))))................... ( -9.92 = -9.96 + 0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:03:44 2011