| Sequence ID | dm3.chr3L |
|---|---|
| Location | 4,797,267 – 4,797,365 |
| Length | 98 |
| Max. P | 0.993057 |

| Location | 4,797,267 – 4,797,365 |
|---|---|
| Length | 98 |
| Sequences | 12 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 83.63 |
| Shannon entropy | 0.36278 |
| G+C content | 0.40234 |
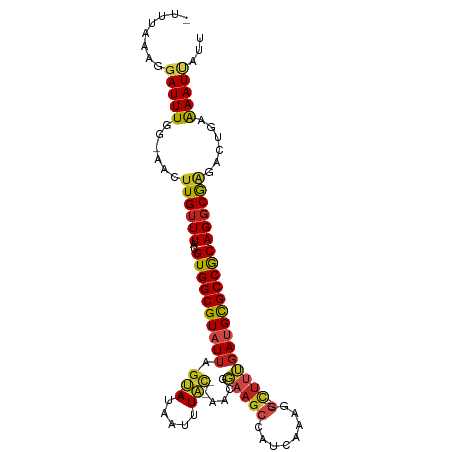
| Mean single sequence MFE | -29.59 |
| Consensus MFE | -18.53 |
| Energy contribution | -18.68 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.59 |
| SVM RNA-class probability | 0.993057 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
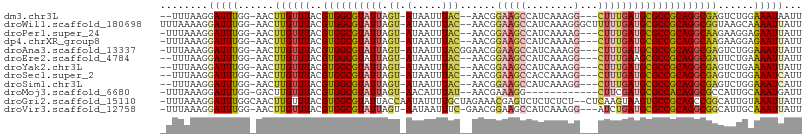
>dm3.chr3L 4797267 98 - 24543557 --UUUAAGGAUUUGG-AACUUGUUUACGUGGCGUAUUAGU-AUAAUUUAC--AACGGAAGCCAUCAAAGG---CUUUGAUGCGCCGCAGGCGAGUCUGGAAAUAAUU --........((..(-(.(((((((..((((((((((.((-(.....)))--....((((((......))---)))))))))))))))))))))))..))....... ( -34.60, z-score = -4.42, R) >droWil1.scaffold_180698 902509 103 + 11422946 UUUAAAAGGAUUUGG-AACUUGUUUACGUGGCGUAUUAGU-AUAAUUUAC--AACGGAAGCCAUCAAAGGGCUUUUUGAUGCGCCGCAGGCGGUAAGCAAAAUUAUU ...............-...(((((((((((((((((((((-(.....)))--...(((((((.......)))))))))))))))))).....))))))))....... ( -30.50, z-score = -2.89, R) >droPer1.super_24 1440232 99 + 1556852 -UUUAAAGGAUUUGG-AACUUGUUUACGUGGCGUAUUAGU-AUAAUUUAC--AACGGAAGCCAUCAAAAG---CUUUGAUGCGCCGCAGGCAAGAAGGAGAAUUAUU -.......(((((..-..(((((((..((((((((((.((-(.....)))--....(((((........)---))))))))))))))))))))).....)))))... ( -27.40, z-score = -2.94, R) >dp4.chrXR_group8 2940500 99 - 9212921 -UUUAAAGGAUUUGG-AACUUGUUUACGUGGCGUAUUAGU-AUAAUUUAC--AACGGAAGCCAUCAAAAG---CUUUGAUGCGCCGCAGGCAAGAAGGAGAAUUAUU -.......(((((..-..(((((((..((((((((((.((-(.....)))--....(((((........)---))))))))))))))))))))).....)))))... ( -27.40, z-score = -2.94, R) >droAna3.scaffold_13337 2885761 101 - 23293914 -UUUAAAGGAUUUGG-AACUUGUUUACGUGGCGUAUUAGU-AUAAUUUACGGAACGGAAGCCAUCAAAGG---CUUUGAUGCGCCGCAGGCGAGUCUGGAAAUUAUU -.........((..(-(.(((((((..((((((((((.((-(.....)))......((((((......))---)))))))))))))))))))))))..))....... ( -34.60, z-score = -4.04, R) >droEre2.scaffold_4784 7513831 98 - 25762168 --UUUAAGGAUUUGG-AACUUGUUUACGUGGCGUAUUAGU-AUAAUUUAC--AACGGAAGCCAUCAAAGG---CUUUGAAGCGCCGCAGGCGAUUCUGAAAAUUAUU --........((..(-((.((((((..(((((((....((-(.....)))--....((((((......))---))))...))))))))))))))))..))....... ( -28.60, z-score = -2.82, R) >droYak2.chr3L 5380291 98 - 24197627 --UUUAAGGAUUUGG-AACUUGUUUACGUGGCGUAUUAGU-AUAAUUUAC--AACGGAAGCCAUCAAAGG---CUUUGAUGCGCCGCAGGCGAGUCUGAAAAUUAUU --........((..(-(.(((((((..((((((((((.((-(.....)))--....((((((......))---)))))))))))))))))))))))..))....... ( -34.20, z-score = -4.39, R) >droSec1.super_2 4783504 98 - 7591821 --UUUAAGGAUUUGG-AACUUGUUUACGUGGCGUAUUAGU-AUAAUUUAC--AACGGAAGCCACCAAAGG---CUUUGAUGCGCCGCAGGCGAGUCUGGAAAUCAUU --........((..(-(.(((((((..((((((((((.((-(.....)))--....((((((......))---)))))))))))))))))))))))..))....... ( -34.60, z-score = -4.03, R) >droSim1.chr3L 4332426 98 - 22553184 --UUUAAGGAUUUGG-AACUUGUUUACGUGGCGUAUUAGU-AUAAUUUAC--AACGGAAGCCAUCAAAGG---CUUUGAUGCGCCGCAGGCGAGUCUGGAAAUCAUU --........((..(-(.(((((((..((((((((((.((-(.....)))--....((((((......))---)))))))))))))))))))))))..))....... ( -34.60, z-score = -4.24, R) >droMoj3.scaffold_6680 1579447 90 - 24764193 -UUUAAAGGAUUUGG-GACUUGUUUACGUGGCGUAUUAGU-AACAUUUAU--AACGAAAGG------------CUUCGAUGCGCCACAGGCGCCAUUGCAAAUGAUU -........(((((.-((..(((((..(((((((((((((-....(((..--....))).)------------))..)))))))))))))))...)).))))).... ( -20.50, z-score = -0.47, R) >droGri2.scaffold_15110 23595475 104 + 24565398 -UUUAAAGGAUUUGGCAACUUGUUUACGUGGCGUAUUACCAAUAUUUUGCUAGAAACGAGUCUCUCUCU--CUCAAGUAAGUGCCGCAGCCGGCAUUGUAAAUUAUU -......(..((((((((..((((((((...)))).....))))..))))))))..)(((.....))).--........(((((((....))))))).......... ( -21.00, z-score = 0.11, R) >droVir3.scaffold_12758 486943 100 - 972128 -UUUAAAGGAUUUGG-AACUUGUUUACGUGGCGUAUUAGU-AAUAAUUUC-GAACGGAAGCCAUCAAAGG---AUCUGAUGCGCCGCAGGCGGCAUUGCAAAUUAUU -.......((((((.-((..((((..(((((((((((((.-.....((((-....)))).((......))---..)))))))))))).)..)))))).))))))... ( -27.10, z-score = -1.56, R) >consensus _UUUAAAGGAUUUGG_AACUUGUUUACGUGGCGUAUUAGU_AUAAUUUAC__AACGGAAGCCAUCAAAGG___CUUUGAUGCGCCGCAGGCGAGACUGAAAAUUAUU ........(((((......((((((..((((((((((((...............(....)...............))))))))))))))))))......)))))... (-18.53 = -18.68 + 0.14)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:03:40 2011