
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 4,788,817 – 4,788,909 |
| Length | 92 |
| Max. P | 0.741535 |

| Location | 4,788,817 – 4,788,909 |
|---|---|
| Length | 92 |
| Sequences | 8 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 61.65 |
| Shannon entropy | 0.74399 |
| G+C content | 0.45392 |
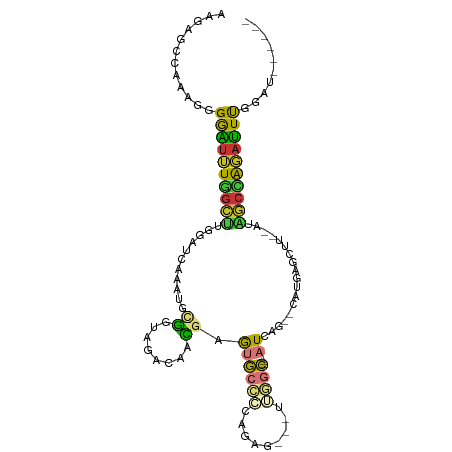
| Mean single sequence MFE | -26.70 |
| Consensus MFE | -7.89 |
| Energy contribution | -7.16 |
| Covariance contribution | -0.73 |
| Combinations/Pair | 2.06 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.741535 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
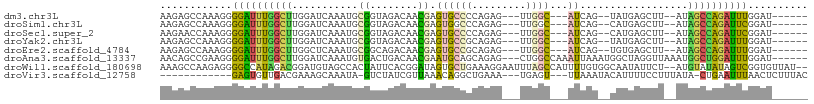
>dm3.chr3L 4788817 92 - 24543557 AAGAGCCAAAGGGGAUUUGGCUUGGAUCAAAUGCGGUAGACAACGAGUGCCCCAGAG---UUGGC---AUCAG--UAUGAGCUU--AUAGCCAGAUUUGGAU------ ..((((((((.....))))))))...........((((.........))))(((((.---(((((---((.((--(....))).--)).))))).)))))..------ ( -25.30, z-score = -0.86, R) >droSim1.chr3L 4322829 92 - 22553184 AAGAGCCAAAGGGGAUUUGGCUUGGAUCAAAUGCGGUAGACAACGAGUGGCCCAGAG---UUGGC---AUCAG--CAUGAGCUU--AUAGCCAGAUUCGGAU------ ..((((((((.....))))))))((..((....((........))..))..)).(((---(((((---((.((--(....))).--)).)))).))))....------ ( -27.70, z-score = -1.34, R) >droSec1.super_2 4775055 92 - 7591821 AAGAACCAAAGGGGAUUUGGCUUGGAUCAAAUGCGGUAGACAACGAGUGCCCCAGAG---UUGGC---AUCAG--CAUGAGCUU--AUAGCCAGAUUCGGAU------ .....((...(((((((((......(((......)))......))))).)))).(((---(((((---((.((--(....))).--)).)))).))))))..------ ( -26.50, z-score = -1.19, R) >droYak2.chr3L 5371716 92 - 24197627 AAGAGCCAAAGGGGAUUUGGCUUGGAUCAAAUGCGGUAGACAACGAGUGCCGCAGAG---UUGGC---AUCAG--UAUGAGCUU--AUAGCCAGAUUUGGAU------ ..((((((((.....))))))))(((((...(((((((.........)))))))...---.((((---((.((--(....))).--)).)))))))))....------ ( -28.60, z-score = -2.22, R) >droEre2.scaffold_4784 7505350 92 - 25762168 AAGAGCCAAAGGGGAUUUGGCUUGGCUCAAAUGCGGCAGACAACGAGUGCCGCAGAG---UUGGC---AUCAG--UGUGAGCUU--AUAGCCAGAUUUGGAU------ ..((((((((.....))))))))((((....(((((((.........)))))))(((---((.((---(....--))).)))))--..))))..........------ ( -33.80, z-score = -2.54, R) >droAna3.scaffold_13337 2876869 99 - 23293914 AACAGCCGAAGGGGAUUUGGCUUGGAUCAAAUGUGACUGACAACGAAUGCAGCAGAG---CUGGCCAAAUUAAAUGGCUAGGUUAAAUGGCUGGAUUUGGAU------ ..((((((((.....)))))).)).(((...(((.....))).(((((.((((.((.---(((((((.......))))))).)).....)))).))))))))------ ( -30.70, z-score = -2.94, R) >droWil1.scaffold_180698 892800 104 + 11422946 AAAGCCAAGAGGGGCCAUAGACGGAUGUAGCCACUAUUCACGGAUAGUGCUGAAAGGAAUUUAGCCAUUUUGUGGCAAUAUUCU--AUGUAUAUAGUCGGUGUUAU-- ...(((......)))(((.(((..(((((((.((((((....)))))))).....(((((...(((((...)))))...)))))--...))))).))).)))....-- ( -27.30, z-score = -1.10, R) >droVir3.scaffold_12758 477041 88 - 972128 ------------GAGUGUUGACGAAAGCAAAUA-GUCUAUCGUUAAACAGGCUGAAA---UGAGU---UUAAAUACAUUUUCCUUUAUA-CUGAAUUUAACUCUUUAC ------------((((.(((((((.(((.....-).)).)))))))..(((..((((---((.((---....)).))))))))).....-.........))))..... ( -13.70, z-score = -0.45, R) >consensus AAGAGCCAAAGGGGAUUUGGCUUGGAUCAAAUGCGGUAGACAACGAGUGCCCCAGAG___UUGGC___AUCAG__CAUGAGCUU__AUAGCCAGAUUUGGAU______ ............((((((((((...........((........))...((((.........)))).......................)))))))))).......... ( -7.89 = -7.16 + -0.73)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:03:39 2011