| Sequence ID | dm3.chr3L |
|---|---|
| Location | 4,776,472 – 4,776,573 |
| Length | 101 |
| Max. P | 0.689752 |

| Location | 4,776,472 – 4,776,573 |
|---|---|
| Length | 101 |
| Sequences | 8 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 69.81 |
| Shannon entropy | 0.56711 |
| G+C content | 0.41760 |
| Mean single sequence MFE | -23.49 |
| Consensus MFE | -9.14 |
| Energy contribution | -10.18 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.689752 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 4776472 101 - 24543557 CUGACUUUUUUUUCACUUUGC---------UCGCCUUUGUUG--CCGAAUC--AAAGUGACUUUUGCUUGAAGUCGCCGACGACAACACAACAUUUUGUAAUUUUCGUCAUAUU ..((((((....((((((((.---------(((.(......)--.)))..)--))))))).........))))))...(((((....((((....)))).....)))))..... ( -22.22, z-score = -2.27, R) >droEre2.scaffold_4784 7492863 100 - 25762168 CUGGC-UUUUCUUCACUUGGC---------UCGCCUUUGUGG--CCGAAUC--AAAGUGACUUUUGCUUGAAGUCACCGACGACGACACAACAUUUUGUAAUUUUCGUCAUAUU .....-..........(((((---------.(((....))))--))))...--...((((((((.....)))))))).(((((.((.((((....))))..)).)))))..... ( -24.80, z-score = -1.88, R) >droYak2.chr3L 5358812 109 - 24197627 UUGGC-UUUUCUUCACUUUGUGGUCCUUUGUGGUCUUUGUGG--CCGAAUC--AAAGUGACUUUUGCUUGAAGUUGCCGACGACGACACAACAUUUUGUAAUUUUCGUCAUAUU ..(((-....(((((......((((((((((((((.....))--)))...)--)))).))))......)))))..)))(((((.((.((((....))))..)).)))))..... ( -31.50, z-score = -2.82, R) >droSim1.chr3L 4308834 100 - 22553184 CUGGC-UUUUCUUCACUUUGC---------UCGCCUUUGUGG--CCGAAAC--AAAGUGACUUUUGCUUGAAGUCGCCGACAACGACACAACAUUUUGUAAUUUUCGUCAUAUU ..(((-........((((((.---------(((((.....))--.)))..)--)))))((((((.....)))))))))(((......((((....)))).......)))..... ( -23.22, z-score = -1.55, R) >droSec1.super_2 4762674 100 - 7591821 CAGGC-UUUUCUUCACUUUGC---------UCGCCUUUGUGG--CCGAAAC--AAAGUGACUUUUGCUUGAAGUCGCCGACGACGACACAACAUUUUGUAAUUUUCGUCAUAUU ..(((-........((((((.---------(((((.....))--.)))..)--)))))((((((.....)))))))))(((((.((.((((....))))..)).)))))..... ( -28.00, z-score = -2.74, R) >droAna3.scaffold_13337 2864246 91 - 23293914 ----------UUUCACUCGCC--------GGCUCCUUUAUGG--CCAAAUCAAAAAGUGACUUUUGCUUGAAGUUGCCGACGAC---ACAGCUUUUUGUAAUUUUCGUCAUAUU ----------...........--------(((..(((((.((--(.((((((.....))).))).))))))))..)))(((((.---...((.....)).....)))))..... ( -19.40, z-score = -1.21, R) >dp4.chrXR_group8 2917789 83 - 9212921 -----------------CUGCG-------AGUCCUUUUGUGGACCGGAGAAUCAAAGUGACUUCUGCUUGAAGUUGCCGACAAC-------UUUUCUGUAAUUUUCGUCAGACU -----------------(((((-------(((((......))))(((((((.....(..(((((.....)))))..).......-------)))))))......))).)))... ( -24.20, z-score = -1.90, R) >droGri2.scaffold_15110 23575482 86 + 24565398 ---------------UUUUG---------GGCGUGUUUUUUGCGCCAAAUCAAAAAACUGACGCUGCUUGAAGUU-UCGCCGAC---AUGUUGCCUUCCAAUUUUCAUCAUCAU ---------------.....---------(((..((((((((........)))))))).((.(((......))).-)))))((.---..((((.....))))..))........ ( -14.60, z-score = 0.91, R) >consensus CUGGC_UUUUCUUCACUUUGC_________UCGCCUUUGUGG__CCGAAUC__AAAGUGACUUUUGCUUGAAGUCGCCGACGACGACACAACAUUUUGUAAUUUUCGUCAUAUU ........................................................((((((((.....)))))))).(((((....((((....)))).....)))))..... ( -9.14 = -10.18 + 1.03)

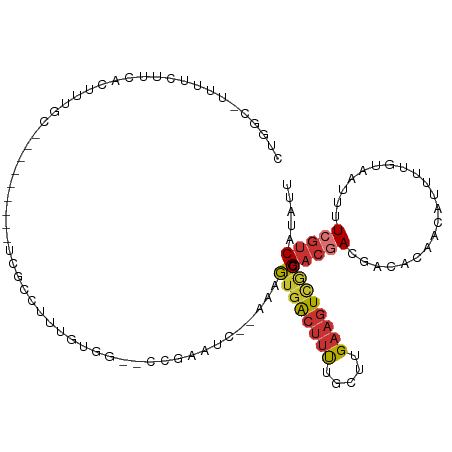

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:03:36 2011