| Sequence ID | dm3.chr2L |
|---|---|
| Location | 4,325,880 – 4,326,003 |
| Length | 123 |
| Max. P | 0.983054 |

| Location | 4,325,880 – 4,325,996 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 124 |
| Reading direction | forward |
| Mean pairwise identity | 76.19 |
| Shannon entropy | 0.38446 |
| G+C content | 0.50703 |
| Mean single sequence MFE | -35.08 |
| Consensus MFE | -18.87 |
| Energy contribution | -18.82 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.589918 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
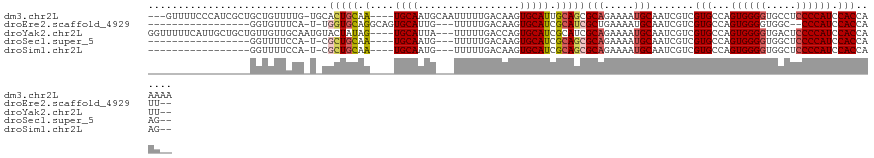
>dm3.chr2L 4325880 116 + 23011544 ---GUUUUCCCAUCGCUGCUGUUUUG-UGCACUGCAA----UGCAAUGCAAUUUUUGACAAGUGCAUUGCAGCGCAGAAAAUGCAAUCGUCGUGCCAGUGGGGUGCCUCCCCAUCCACCAAAAA ---.............(((.(((((.-(((.((((((----((((.((((.....)).))..)))))))))).))).))))))))......(((...((((((.....)))))).)))...... ( -39.90, z-score = -2.38, R) >droEre2.scaffold_4929 4402807 98 + 26641161 -----------------GGUGUUUCA-U-UGGUGCAGGCAGUGCAUUG---UUUUUGACAAGUGCAUCGCAUCGCUGAAAAUGCAAUCGUCGUGCCAGUGGGGUGGC--CCCAUCCACCAUU-- -----------------(((....((-(-(((..(.(((.((((((((---((...))).))))))).((((........))))....))))..))))))(((((..--..))))))))...-- ( -35.90, z-score = -1.45, R) >droYak2.chr2L 4357313 115 + 22324452 GGUUUUUCAUUGCUGCUGUUGUUGCAAUGUACUAUAG----UGCAUUA---UUUUUGACCAGUGCAUCGCAUCGCAGAAAAUGCAAUCGUCGUGCCAGUGGGGUGACUCCCCAUCCACCAUU-- (((....((((((..((((...(((.(((((((...(----(.((...---....)))).))))))).)))..)))).....))))).)....))).((((((.....))))))........-- ( -32.80, z-score = -1.07, R) >droSec1.super_5 2423457 96 + 5866729 -----------------GGUUUUCCA-U-CGCUGCAA----UGCAAUG---UUUUUGACAAGUGCAUCGCAGCGCAGAAAAUGCAAUCGUCGUGCCAGUGGGGUGGCUCCCCAUCCACCAAG-- -----------------.((((((..-.-((((((.(----((((.((---((...))))..))))).))))))..)))))).........(((...((((((.....)))))).)))....-- ( -33.40, z-score = -2.00, R) >droSim1.chr2L 4248082 96 + 22036055 -----------------GGUUUUCCA-U-CGCUGCAA----UGCAAUG---UUUUUGACAAGUGCAUCGCAGCGCAGAAAAUGCAAUCGUCGUGCCAGUGGGGUGGCUCCCCAUCCACCAAG-- -----------------.((((((..-.-((((((.(----((((.((---((...))))..))))).))))))..)))))).........(((...((((((.....)))))).)))....-- ( -33.40, z-score = -2.00, R) >consensus _________________GGUGUUCCA_U_CGCUGCAA____UGCAAUG___UUUUUGACAAGUGCAUCGCAGCGCAGAAAAUGCAAUCGUCGUGCCAGUGGGGUGGCUCCCCAUCCACCAAG__ ............................(((.((((.....)))).))).......(((...(((((..(......)...)))))...)))(((...((((((.....)))))).)))...... (-18.87 = -18.82 + -0.05)
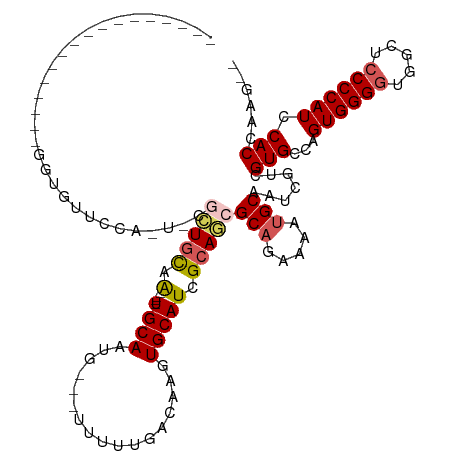
| Location | 4,325,880 – 4,325,996 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 124 |
| Reading direction | reverse |
| Mean pairwise identity | 76.19 |
| Shannon entropy | 0.38446 |
| G+C content | 0.50703 |
| Mean single sequence MFE | -36.40 |
| Consensus MFE | -21.66 |
| Energy contribution | -21.94 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.983054 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 4325880 116 - 23011544 UUUUUGGUGGAUGGGGAGGCACCCCACUGGCACGACGAUUGCAUUUUCUGCGCUGCAAUGCACUUGUCAAAAAUUGCAUUGCA----UUGCAGUGCA-CAAAACAGCAGCGAUGGGAAAAC--- ....((.(((.(((((.....)))))))).))((.((.((((.((((.((((((((((((((..(((........))).))))----))))))))))-.))))..)))))).)).......--- ( -43.30, z-score = -2.85, R) >droEre2.scaffold_4929 4402807 98 - 26641161 --AAUGGUGGAUGGG--GCCACCCCACUGGCACGACGAUUGCAUUUUCAGCGAUGCGAUGCACUUGUCAAAAA---CAAUGCACUGCCUGCACCA-A-UGAAACACC----------------- --..(((((..((((--(...)))))..((((.....((((((((......))))))))(((.((((.....)---))))))..))))..)))))-.-.........----------------- ( -30.60, z-score = -1.64, R) >droYak2.chr2L 4357313 115 - 22324452 --AAUGGUGGAUGGGGAGUCACCCCACUGGCACGACGAUUGCAUUUUCUGCGAUGCGAUGCACUGGUCAAAAA---UAAUGCA----CUAUAGUACAUUGCAACAACAGCAGCAAUGAAAAACC --.((((((.((((((.....))))((..(..((.((((((((.....)))))).)).))..)..))......---..)).))----))))....((((((..........))))))....... ( -30.90, z-score = -0.99, R) >droSec1.super_5 2423457 96 - 5866729 --CUUGGUGGAUGGGGAGCCACCCCACUGGCACGACGAUUGCAUUUUCUGCGCUGCGAUGCACUUGUCAAAAA---CAUUGCA----UUGCAGCG-A-UGGAAAACC----------------- --...(((((.(((....)))..))))).(((.......))).(((((((((((((((((((..(((.....)---)).))))----))))))))-.-)))))))..----------------- ( -38.60, z-score = -3.71, R) >droSim1.chr2L 4248082 96 - 22036055 --CUUGGUGGAUGGGGAGCCACCCCACUGGCACGACGAUUGCAUUUUCUGCGCUGCGAUGCACUUGUCAAAAA---CAUUGCA----UUGCAGCG-A-UGGAAAACC----------------- --...(((((.(((....)))..))))).(((.......))).(((((((((((((((((((..(((.....)---)).))))----))))))))-.-)))))))..----------------- ( -38.60, z-score = -3.71, R) >consensus __AUUGGUGGAUGGGGAGCCACCCCACUGGCACGACGAUUGCAUUUUCUGCGCUGCGAUGCACUUGUCAAAAA___CAUUGCA____UUGCAGCG_A_UGAAACACC_________________ ....((.(((.(((((.....)))))))).)).(((((.((((((....((...)))))))).)))))...........((((.....))))................................ (-21.66 = -21.94 + 0.28)
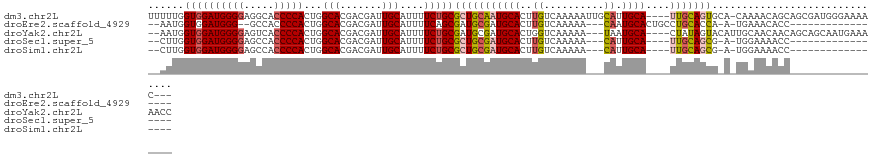
| Location | 4,325,887 – 4,326,003 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.59 |
| Shannon entropy | 0.37283 |
| G+C content | 0.53780 |
| Mean single sequence MFE | -35.82 |
| Consensus MFE | -25.75 |
| Energy contribution | -26.87 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.774445 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 4325887 116 - 23011544 CCAUGGGUUUUUGGUGGAUGGGGAGGCACCCCACUGGCACGACGAUUGCAUUUUCUGCGCUGCAAUGCACUUGUCAAAAAUUGCAUUGCAUUGCAGUGCACAA-AACAGCAGCGAUG--- .(((.((((..((.(((.(((((.....)))))))).)).)))..((((.((((.((((((((((((((..(((........))).)))))))))))))).))-))..))))).)))--- ( -43.10, z-score = -1.79, R) >droEre2.scaffold_4929 4402814 98 - 26641161 CCACGGG--AAUGGUGGAUGGG--GCCACCCCACUGGCACGACGAUUGCAUUUUCAGCGAUGCGAUGCACUUGUCAAAAA---CAAUGCACUGC-CUGCACCA-AUG------------- .......--..(((((..((((--(...)))))..((((.....((((((((......))))))))(((.((((.....)---))))))..)))-)..)))))-...------------- ( -30.60, z-score = -0.70, R) >droYak2.chr2L 4357320 115 - 22324452 CCGCGGG--AAUGGUGGAUGGGGAGUCACCCCACUGGCACGACGAUUGCAUUUUCUGCGAUGCGAUGCACUGGUCAAAAA---UAAUGCACUAUAGUACAUUGCAACAACAGCAGCAAUG .((((((--((....((.(((((.....))))))).(((.......)))..)))))))).(((((((.((((((((....---...)).)))..))).)))))))............... ( -30.80, z-score = 0.27, R) >droSec1.super_5 2423464 96 - 5866729 CCACGGG--CUUGGUGGAUGGGGAGCCACCCCACUGGCACGACGAUUGCAUUUUCUGCGCUGCGAUGCACUUGUCAAAAA---CAUUGCAUUGCAGCG-AUG------------------ ...((.(--((.(((((.(((....)))..)))))))).))......(((.....)))(((((((((((..(((.....)---)).))))))))))).-...------------------ ( -37.30, z-score = -2.43, R) >droSim1.chr2L 4248089 96 - 22036055 CCACGGG--CUUGGUGGAUGGGGAGCCACCCCACUGGCACGACGAUUGCAUUUUCUGCGCUGCGAUGCACUUGUCAAAAA---CAUUGCAUUGCAGCG-AUG------------------ ...((.(--((.(((((.(((....)))..)))))))).))......(((.....)))(((((((((((..(((.....)---)).))))))))))).-...------------------ ( -37.30, z-score = -2.43, R) >consensus CCACGGG__AUUGGUGGAUGGGGAGCCACCCCACUGGCACGACGAUUGCAUUUUCUGCGCUGCGAUGCACUUGUCAAAAA___CAUUGCAUUGCAGUGCAUG__A_______________ ((((.(.....).)))).(((((.....)))))...(((.......))).......(((((((((((((.................)))))))))))))..................... (-25.75 = -26.87 + 1.12)

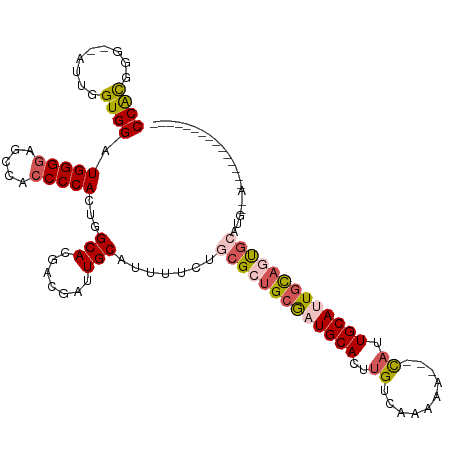
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:16:09 2011