
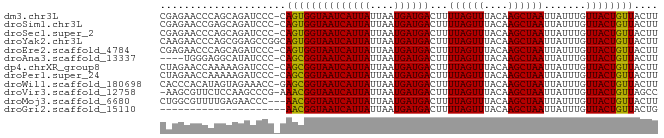
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 4,735,030 – 4,735,112 |
| Length | 82 |
| Max. P | 0.617371 |

| Location | 4,735,030 – 4,735,112 |
|---|---|
| Length | 82 |
| Sequences | 12 |
| Columns | 83 |
| Reading direction | forward |
| Mean pairwise identity | 84.78 |
| Shannon entropy | 0.36647 |
| G+C content | 0.32531 |
| Mean single sequence MFE | -13.12 |
| Consensus MFE | -10.60 |
| Energy contribution | -10.17 |
| Covariance contribution | -0.43 |
| Combinations/Pair | 1.10 |
| Mean z-score | -0.95 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.617371 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
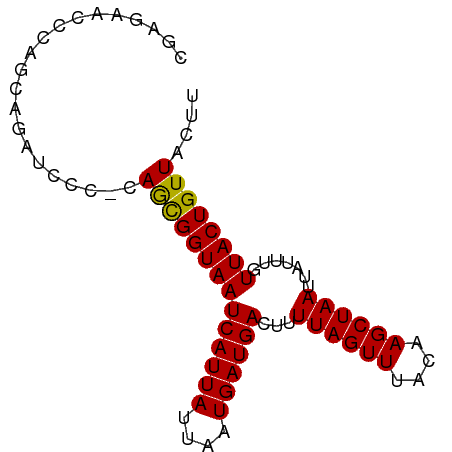
>dm3.chr3L 4735030 82 + 24543557 CGAGAACCCAGCAGAUCCC-CAGUGGUAAUCAUUAUUAAUGAUGACUUUUAGUUUACAAGCUAAUUAUUUGUUACUGUUACUU .((((((..(((((((...-....(((.(((((.....))))).))).((((((....))))))..)))))))...))).))) ( -11.50, z-score = -0.28, R) >droSim1.chr3L 4267611 82 + 22553184 CGAGAACCGAGCAGAUCCC-CAGUGGUAAUCAUUAUUAAUGAUGACUUUUAGUUUACAAGCUAAUUAUUUGUUACUGUUACUU .((((((.((((((((...-....(((.(((((.....))))).))).((((((....))))))..))))))).).))).))) ( -12.70, z-score = -0.57, R) >droSec1.super_2 4718789 82 + 7591821 CGAGAACCCAGCAGAUCCC-CAGUGGUAAUCAUUAUUAAUGAUGACUUUUAGUUUACAAGCUAAUUAUUUGUUACUGUUACUU .((((((..(((((((...-....(((.(((((.....))))).))).((((((....))))))..)))))))...))).))) ( -11.50, z-score = -0.28, R) >droYak2.chr3L 5317576 83 + 24197627 CAAGAACCCAGCGGAGCCGGCAGUGGUAAUCAUUAUUAAUGAUGACUUUUAGUUUACAAGCUAAUUAUUUGUUACUGUUACUU .............(((..((((((..(((((((((....))))))...((((((....))))))....)))..)))))).))) ( -14.30, z-score = 0.06, R) >droEre2.scaffold_4784 7451157 82 + 25762168 CGAGAACCCAGCAGAUCCC-CAGUGGUAAUCAUUAUUAAUGAUGACUUUUAGUUUACAAGCUAAUUAUUUGUUACUGUUACUU .((((((..(((((((...-....(((.(((((.....))))).))).((((((....))))))..)))))))...))).))) ( -11.50, z-score = -0.28, R) >droAna3.scaffold_13337 2824154 78 + 23293914 ----UGGGAGGCAUAUCCC-CAGCGGUAAUCAUUAUUAAUGAUGACUUUUAGUUUACAAGCUAAUUAUUUGUUACUGUUACUU ----.((((......))))-.((((((((((((((....))))))...((((((....)))))).......)))))))).... ( -18.00, z-score = -1.89, R) >dp4.chrXR_group8 2862733 82 + 9212921 CUAGAACCAAAAAGAUCCC-CAGCGGUAAUCAUUAUUAAUGAUGACUUUUAGUUUACAAGCUAAUUAUUUGUUACUGUUACUU ...................-.((((((((((((((....))))))...((((((....)))))).......)))))))).... ( -12.50, z-score = -1.11, R) >droPer1.super_24 1366780 82 - 1556852 CUAGAACCAAAAAGAUCCC-CAGCGGUAAUCAUUAUUAAUGAUGACUUUUAGUUUACAAGCUAAUUAUUUGUUACUGUUACUU ...................-.((((((((((((((....))))))...((((((....)))))).......)))))))).... ( -12.50, z-score = -1.11, R) >droWil1.scaffold_180698 837809 82 - 11422946 CACCCACAUAGUAGAAACC-GAGCGGUAAUCAUUAUUAAUGAUGACUUUUAGUUUACAAGCUAAUUAUUUGUUACUGUUACUU ...................-.((((((((((((((....))))))...((((((....)))))).......)))))))).... ( -12.60, z-score = -0.84, R) >droVir3.scaffold_12758 401578 81 + 972128 -AAGCGUUCUCCAAGCCCG-AAACGGUAAUCAUUAUUAAUGAUGACUUUUAGUUUACAAGCUAAUUAUUUGUUACUGUUAGCC -..(((((.....)))...-.((((((((((((((....))))))...((((((....)))))).......)))))))).)). ( -13.00, z-score = -0.75, R) >droMoj3.scaffold_6680 1501417 80 + 24764193 CUGGCGUUUUGAGAACCC---AACGGUAAUCAUUAUUAAUGAUGACUUUUAGUUUACAAGCUAAUUAUUUGUUACUGUUACUU ..((.((((...))))))---((((((((((((((....))))))...((((((....)))))).......)))))))).... ( -15.20, z-score = -1.70, R) >droGri2.scaffold_15110 23531060 62 - 24565398 ---------------------AACGGUAAUCAUUAUUAAUGAUGACUUUUAGUUUACAAGCUAAUUAUUUGUUACUGUUACUG ---------------------((((((((((((((....))))))...((((((....)))))).......)))))))).... ( -12.20, z-score = -2.64, R) >consensus CGAGAACCCAGCAGAUCCC_CAGCGGUAAUCAUUAUUAAUGAUGACUUUUAGUUUACAAGCUAAUUAUUUGUUACUGUUACUU .....................((((((((((((((....))))))...((((((....)))))).......)))))))).... (-10.60 = -10.17 + -0.43)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:03:32 2011