
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 4,693,939 – 4,694,039 |
| Length | 100 |
| Max. P | 0.783869 |

| Location | 4,693,939 – 4,694,039 |
|---|---|
| Length | 100 |
| Sequences | 10 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 74.41 |
| Shannon entropy | 0.49942 |
| G+C content | 0.40074 |
| Mean single sequence MFE | -25.29 |
| Consensus MFE | -11.12 |
| Energy contribution | -11.66 |
| Covariance contribution | 0.54 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.783869 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 4693939 100 - 24543557 UGUGGCAAUUACACAA--AAAUGUCAAUUGUCCGCGG----AUUUGUUGUAGGAGUGGAAAUUCCUGUUCU--GUAAGCCUGUUUUUGGC-UCUGUUUUUGCUAGCUAG---- ..((((((..(((((.--......((((.(((....)----))..))))(((((((....)))))))...)--)).((((.......)))-)..))..)))))).....---- ( -25.80, z-score = -1.41, R) >droSim1.chr3L 4225824 100 - 22553184 UCAGGCAAUUACACAA--AAAUGUCAAUUGUCCGCGG----AUUUGUUGUAGGAGUGGAAAUUCCUGUUCU--GUAAGCCUGUUUUUGGC-UCUGUUUUUGCUAGCUAG---- .(((((..(((((...--......((((.(((....)----))..))))(((((((....)))))))...)--)))))))))....((((-(..((....)).))))).---- ( -26.90, z-score = -1.67, R) >droSec1.super_2 4675907 88 - 7591821 UCUGGCAAUUACACAA--AAAUGUCAAUUGUCCGCGG----AUUUGUUGUAGGAGUGGAAAUUCCUGUUCU--GUAAGCCUGUUUUUGGUAGCUAG----------------- .(((((..(((((...--......((((.(((....)----))..))))(((((((....)))))))...)--))))(((.......))).)))))----------------- ( -22.90, z-score = -1.70, R) >droYak2.chr3L 5275370 104 - 24197627 UCUGGCAAUUACACAA--AAAUGUCAAUUGUCCGCGG----AUUUGUUGUAGGAGUGGAAAUUCCUGUUCU--GCAAGCCUGUUUUUGGC-UCUGUUUUUGCCAGCUAGAGUU .(((((((..(((...--...............((((----(.......(((((((....))))))).)))--)).((((.......)))-).)))..)))))))........ ( -31.60, z-score = -2.14, R) >droEre2.scaffold_4784 7408586 100 - 25762168 UCUGGCAAUUACACAA--AAAUGUCAAUUGUCCGCGG----AUUUGUUGUAGGAGUGGAAAUUCCUGUUCU--GUAAGCCUGUUUUUGGC-UCUGUUUUUGCCAGCUAG---- .(((((((..(((((.--......((((.(((....)----))..))))(((((((....)))))))...)--)).((((.......)))-)..))..)))))))....---- ( -30.20, z-score = -2.72, R) >droAna3.scaffold_13337 2784137 98 - 23293914 UCUUGCAAUUACACAA--AAAUGUCAAUUGU-UGGGG----CUUUGUUGUAGGAGUGGAAAUUCCUGUUCU--GUAAGCCUGUUUUUGGC-UCUGUUUUUGCGA-CUAG---- ..((((((..(((...--....(((((....-..(((----(((.....(((((((....)))))))....--..))))))....)))))-..)))..))))))-....---- ( -26.70, z-score = -1.95, R) >droWil1.scaffold_180698 797782 93 + 11422946 UGUGAACACGGUAUAA--AAAUGUCAAUUGUCGGCUAUGAUGCUUGUUGUAGGAGUGGAAAUUCCUGUUCUCAAAAAGCCUGUUUUUACGCUGUG------------------ ......(((((..(((--((((.......((((....))))((((.((((((((((....)))))))....))).))))..)))))))..)))))------------------ ( -25.10, z-score = -2.65, R) >droPer1.super_24 1316064 86 + 1556852 UUUGGCAAUUACACAA--AAAUGUCAAUUGUCGGUUG----GUUUGUUGUAGGAGUGGAAAUUCCUGUUCU--GUAAGCCUGUUUUUUUU-UUGG------------------ .((((((((((((...--...))).)))))))))..(----(((((..((((((((....)))))))..).--.))))))..........-....------------------ ( -21.40, z-score = -2.11, R) >dp4.chrXR_group8 2814683 87 - 9212921 UUUGGCAAUUACACAA--AAAUGUCAAUUGUCGGUUG----GUUUGUUGUAGGAGUGGAAAUUCCUGUUCU--GUAAGCCUGUUUUUUUU-UUUGG----------------- .((((((((((((...--...))).)))))))))..(----(((((..((((((((....)))))))..).--.))))))..........-.....----------------- ( -21.40, z-score = -2.23, R) >droGri2.scaffold_15110 23480676 97 + 24565398 UCUGCAUAUUGUCUGAGUGUAUUUUUGCUAGUUGCUGUUGUUGUUGUUGUAGGAGUGGAAAUGCCUGUUCU--UAAAGCCUGU--UUGGUCUGGGCCAGAA------------ ...........((((((.(((((((..((..((((.............)))).))..))))))))).....--....((((..--.......)))))))).------------ ( -20.92, z-score = -0.15, R) >consensus UCUGGCAAUUACACAA__AAAUGUCAAUUGUCCGCGG____AUUUGUUGUAGGAGUGGAAAUUCCUGUUCU__GUAAGCCUGUUUUUGGC_UCUGUUUUUG____________ ...((((((((((........))).))))))).................(((((((....))))))).............................................. (-11.12 = -11.66 + 0.54)
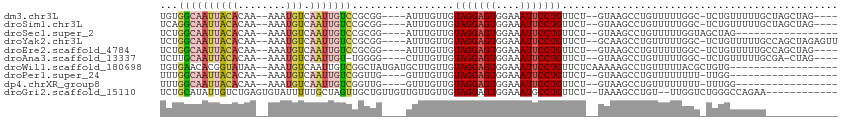
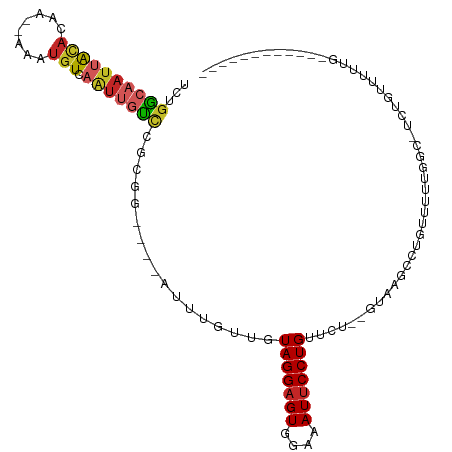

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:03:28 2011