
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 4,677,987 – 4,678,107 |
| Length | 120 |
| Max. P | 0.969807 |

| Location | 4,677,987 – 4,678,107 |
|---|---|
| Length | 120 |
| Sequences | 10 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.23 |
| Shannon entropy | 0.64537 |
| G+C content | 0.54152 |
| Mean single sequence MFE | -43.78 |
| Consensus MFE | -17.18 |
| Energy contribution | -18.15 |
| Covariance contribution | 0.97 |
| Combinations/Pair | 1.58 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.969807 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
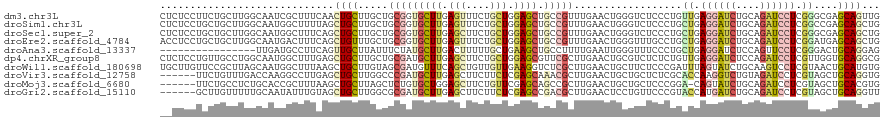
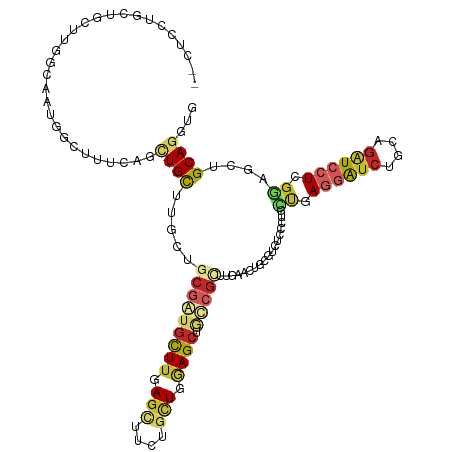
>dm3.chr3L 4677987 120 - 24543557 CUCUCCUUCUGCUUGGCAAUCGCUUUCAACUGCUUGCUGCGGUGCUUGAGUUUCUGCUGGAGCUGCCGUUUGAACUGGGUCUCCCUGUUGAGGAUCUGCAGAUCCUCGGGCGAGCAGUUG ..............(((....)))..(((((((((((((((((((((.(((....))).)))).)))))...(((.(((...))).)))(((((((....))))))).)))))))))))) ( -50.80, z-score = -3.34, R) >droSim1.chr3L 4207753 120 - 22553184 CUCUCCUGCUGCUUGGCAAUGGCUUUUAGCUGCUUGCUGCGGUGCUUGAGUUUCUGCUGGAGCUGCCGUUUGAACUGGGUCUCCCUGCUGAGGAUCUGCAGAUCCUCGGCCGAGCAGCUG .......((((((((((((((((.....(((((.....)))))((((.(((....))).)))).))))))......(((...)))...((((((((....)))))))))))))))))).. ( -55.50, z-score = -3.39, R) >droSec1.super_2 4660225 120 - 7591821 CUCUCCUGCUGCUUGGCAAUGGCUUUCAGCUGCUUGCUGCGGUGCUUGAGCUUCUGCUGGAGCUGCCGUUUGAACUGGGUCUCCCUGCUGAGGAUCUGCAGAUCCUCGGGCGAGCAGCUG ....((((((....))))..))....(((((((((((.(((((((((.(((....))).)))).))))).......(((...)))..(((((((((....)))))))))))))))))))) ( -57.00, z-score = -2.97, R) >droEre2.scaffold_4784 7390401 120 - 25762168 ACCUCCUGCUGCUUGGCAAUGACUUUCAGCUGUUUGCUGCGGUGCUUGAGUUUCUGCUGGAGCUGCCGUUUGAACUGGGUUUGCCUGCUGAGGAUCUGCAGAUCCUCGGAUGAGCAGCUG .......((((((((((((...((((((((.....)))(((((((((.(((....))).)))).)))))..)))..))..)))))..(((((((((....)))))))))..))))))).. ( -51.90, z-score = -3.46, R) >droAna3.scaffold_13337 2766432 104 - 23293914 ----------------UUGAUGCCUUCAGUUGCUUAUUUCUAUGCUUGACUUUUUGCUGAAGCUGCCUUUUGAAUUGGGUUUCCCUGCUGAGGAUCUCCAGUUCCUCGGGACUGCAGGAG ----------------......(((.(((((.(..........((((.((.....).).))))........((((((((..((((....).)))..))))))))...).))))).))).. ( -27.20, z-score = 0.02, R) >dp4.chrXR_group8 2797143 120 - 9212921 CUCUCCUGUUGCCUGGCAAUGGCUUUGAGCUGCUUGCUGCGAUGCUUGAGCUUCUGCUGGAGCGUUCGCUUGAACUGCGUCUCUCUGUUGAGGAUCUCCAGAUCCUCGUUGGUGCAGGCG ....(((((...(..((...(((...(((.(((...(.(((((((((.(((....))).))))).))))..)....))).)))...)))(((((((....)))))))))..).))))).. ( -47.90, z-score = -2.35, R) >droWil1.scaffold_180698 782815 120 + 11422946 UGCUUGUUCCGCUUAGCAAUGGCUUUAAGCUGCUUGUAGCGAUGUUUCAGCUGUUGUUGAAGGUCUCGCUUGAACUGCUUCUCCCGAUUUAGUAUCUGCAAGUCCUCGUAACUGCAUGUG (((..(((.((.((((((((((((....((((....))))((....))))))))))))))(((.((.((..((((((..((....))..)))).)).)).)).))))).))).))).... ( -32.50, z-score = -0.90, R) >droVir3.scaffold_12758 320156 114 - 972128 ------UUCUGUUUGACCAAGGCCUUGAGCUGCUUGGCCCGAUGCUUGAGCUUCUUCUCGAGCAAACGCUUGAACUGCUGCUCUCGCACCAAGGUCUGUAGAUCCUCGUAGCUGCAGGUG ------.........(((..(((..((((((((..((((.(.(((..((((..(...((((((....))))))...)..))))..))).)..)))).))))...))))..)))...))). ( -37.00, z-score = -0.49, R) >droMoj3.scaffold_6680 1424540 113 - 24764193 ------UUCUGCCUCUGCACCGCUUUAAGCUGCUUAGCUCUGUGCUGGAGCUUCUGUUCGAGCAGCCGCUUGAACUGCUGCUCCCGGA-CAGUAUCUGCAGAUCCUCGUAGCUGCACGUG ------.........((((..(((.....((((......((((.(.(((((..(.((((((((....)))))))).)..))))).).)-))).....))))........))))))).... ( -39.72, z-score = -1.09, R) >droGri2.scaffold_15110 23466818 114 + 24565398 ------GCUUGUUUUUGCAAUAUUUGUAGCUGCUUGGCGCGAUGCUUGAGCUUCUUCUCGAGCCGACGCUUGAACUCCUGUUCCCGUACCAUGAUCUGCAGAUCCUCGUAGCUGCAGGUU ------((........))...((((((((((((.(((..((..(((((((......)))))))........((((....)))).))..))).((((....))))...)))))))))))). ( -38.30, z-score = -2.44, R) >consensus __CUCCUGCUGCUUGGCAAUGGCUUUCAGCUGCUUGCUGCGAUGCUUGAGCUUCUGCUGGAGCUGCCGCUUGAACUGCGUCUCCCUGCUGAGGAUCUGCAGAUCCUCGGAGCUGCAGGUG .............................((((.....(((((((((.(((....))).)))).)))))..................((.((((((....)))))).))....))))... (-17.18 = -18.15 + 0.97)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:03:27 2011