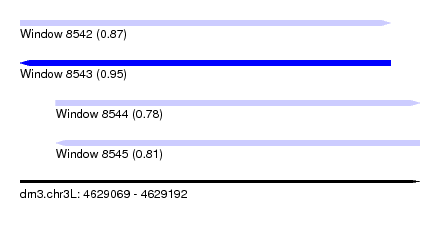
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 4,629,069 – 4,629,192 |
| Length | 123 |
| Max. P | 0.946986 |

| Location | 4,629,069 – 4,629,183 |
|---|---|
| Length | 114 |
| Sequences | 13 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.07 |
| Shannon entropy | 0.46201 |
| G+C content | 0.57373 |
| Mean single sequence MFE | -41.47 |
| Consensus MFE | -20.49 |
| Energy contribution | -21.78 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.871400 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 4629069 114 + 24543557 ------AGGUUCCGGUGCCGGGGCCAAGGGCGCCAAGGAGAACAACAUUUGACGUGGCGGCCGCAAUUGGUUAUAGAAGCCAAUUAAGCGACCGCCGAAGUACAAGCCACCGGAUUUCAC ------.((.(((((((.(((.(((...))).))..............(((((.((((((.(((((((((((.....))))))))..))).))))))..)).)))).)))))))..)).. ( -45.30, z-score = -2.82, R) >droSim1.chr3L 4160668 114 + 22553184 ------AGGUUUUGGUGCCGGGGCCAAGGGCGCCAAGGAGAACAACAUUUGACGUGGCGGCCGCAAUUGGUUAUAGAAGCCAAUUAAGCGACCGCCGAAGUACAAGCCACCGGAUUUCAC ------.((.(((((((.(((.(((...))).))..............(((((.((((((.(((((((((((.....))))))))..))).))))))..)).)))).)))))))..)).. ( -40.40, z-score = -1.67, R) >droSec1.super_2 4611288 114 + 7591821 ------AGGUUCCGGUGCCGGGGCCAAGGGCGCCAAGGAGAACAACAUUUGACGUGGCGGCCGCAAUUGGUUAUAGAAGCCAAUUAAGCGACCGCCGAAGUACAAGCCACCGGAUUUCAC ------.((.(((((((.(((.(((...))).))..............(((((.((((((.(((((((((((.....))))))))..))).))))))..)).)))).)))))))..)).. ( -45.30, z-score = -2.82, R) >droYak2.chr3L 5204211 114 + 24197627 ------AGGUUCCGGUGCCGGGGCCAAGGGCGCCAAGGAGAACAACAUUUGACGUGGCGGCCGCAAUUGGUUAUAGAAGCCAAUUAAGCGACCGCCGAAGUACAAGCCGCCGGAUUUCAC ------.((.(((((((.(((.(((...))).))..............(((((.((((((.(((((((((((.....))))))))..))).))))))..)).)))).)))))))..)).. ( -44.90, z-score = -2.22, R) >droEre2.scaffold_4784 7339133 114 + 25762168 ------GGGUUCCGGUGCCGGGGCCAAGGGCGCCAAGGAGAACAACAUUUGACGUGGCGGCCGCAAUUGGUUAUAGAAGCCAAUUAAGCGACCGCCGAAGUACAAGCCGCCGGAUUUCAC ------(((.(((((((.(((.(((...))).))..............(((((.((((((.(((((((((((.....))))))))..))).))))))..)).)))).))))))).))).. ( -46.20, z-score = -2.32, R) >droAna3.scaffold_13337 2715270 120 + 23293914 AGGAUCCGGAUCCGGAACUGGCGCCAAGGGCACCAAGGAGAACAACAUAUGACGUGGCGGCCGCAAUUGGUUAUAGAAGCCAAUUAAACGACCGCCGAAGUACAAGCCGCCGGACUUCCC .((((((((...(((...(((.(((...))).)))..............((((.((((((.((.((((((((.....))))))))...)).))))))..)).))..))))))))..))). ( -40.40, z-score = -1.60, R) >dp4.chrXR_group8 2741716 114 + 9212921 ------CGGCUCCGGUUCGGGGGCCAAGGCCGCCAAGGAGAACAACAUAUGACGUGGCGGCCGCAAUUGGUUAUAGAAGCCAAUUAAGCGACCGCCGAAGUACAAGCCGCCGGACUUCAC ------(((((..(.(((((.((.(..((((((((.(...............).))))))))((((((((((.....))))))))..))).)).))))).)...)))))........... ( -45.16, z-score = -2.20, R) >droPer1.super_24 1243601 114 - 1556852 ------CGGCUCCGGUUCGGGGGCCAAGGCCGCCAAGGAGAACAACAUAUGACGUGGCGGCCGCAAUUGGUUAUAGAAGCCAAUUAAGCGACCGCCGAAGUACAAGCCGCCGGACUUCAC ------(((((..(.(((((.((.(..((((((((.(...............).))))))))((((((((((.....))))))))..))).)).))))).)...)))))........... ( -45.16, z-score = -2.20, R) >droWil1.scaffold_180698 731153 98 - 11422946 ----------------------ACUAAGGGCGCCAAGGAGAACAACAUAUGACGUGGCGGCCGCAAUUGGUUAUAGAAGCCAAUUAAACGUCCGCCCAAGUACAAGCCGCCAGAUUUUAC ----------------------......((((.(..(.....)......((((..(((((.((.((((((((.....))))))))...)).)))))...)).)).).))))......... ( -26.00, z-score = -1.03, R) >droVir3.scaffold_12758 241215 113 + 972128 -------GGCGCCUCGUCGGCGUCGAAGGGCGCCAAGGAGAACAACAUAUGACGUGGCGGCCGCAAUUGGUUAUAGAAGCCAAUUAAGCGACCGCCCAAAUACAAGCCGCCGGACUUUAC -------((((.(((.(.((((((....)))))).).))).............(.(((((.(((((((((((.....))))))))..))).))))))..........))))......... ( -46.60, z-score = -3.33, R) >droMoj3.scaffold_6680 1358051 113 + 24764193 -------GGUGCCGCCGCUGCAUCGAAGGGCGCCAAGGAGAACAAUAUAUGACGUGGCGGCCGCAAUUGGUUAUAGAAGCCAAUUAAGCGACCGCCCAAAUACAAGCCGCCGGAUUUUAC -------....((((.(((........((((((((.(...............).))))((.(((((((((((.....))))))))..))).)))))).......))).).)))....... ( -34.92, z-score = -0.27, R) >droGri2.scaffold_15110 23409615 113 - 24565398 -------GGCGCCUCCGCGACUAAGGGGGGCGCCAAGGAGAACAACAUUUGACGUGGCGGCCGCAAUUGGUUAUAGAAGCCAAUUAAGCGACCGCCCAAAUAUAAACCGCCCGAUUUUAC -------(((((((((.(......))))))))))..((........(((((....(((((.(((((((((((.....))))))))..))).))))))))))........))......... ( -44.59, z-score = -3.26, R) >anoGam1.chrX 12369530 90 - 22145176 -------------GGCCCCCGGUACACCCGGGCCGGGUGGAGCAGGAGGAGCCGUUGGAGCAGCAGGUGCU----GGAGC------AGCAACGGCAGUAGUAAGCAACGUCAA------- -------------(((((((.((.((((((...))))))..)).)).)).(((((((..(((((....)))----...))------..))))))).............)))..------- ( -34.20, z-score = -0.08, R) >consensus _______GGUUCCGGUGCCGGGGCCAAGGGCGCCAAGGAGAACAACAUAUGACGUGGCGGCCGCAAUUGGUUAUAGAAGCCAAUUAAGCGACCGCCGAAGUACAAGCCGCCGGAUUUCAC .......(((.................(..(.(....).)..)............(((((.(((((((((((.....))))))))..))).)))))............)))......... (-20.49 = -21.78 + 1.28)

| Location | 4,629,069 – 4,629,183 |
|---|---|
| Length | 114 |
| Sequences | 13 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.07 |
| Shannon entropy | 0.46201 |
| G+C content | 0.57373 |
| Mean single sequence MFE | -41.77 |
| Consensus MFE | -24.79 |
| Energy contribution | -25.36 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.946986 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 4629069 114 - 24543557 GUGAAAUCCGGUGGCUUGUACUUCGGCGGUCGCUUAAUUGGCUUCUAUAACCAAUUGCGGCCGCCACGUCAAAUGUUGUUCUCCUUGGCGCCCUUGGCCCCGGCACCGGAACCU------ ......(((((((.(.........(((((((((..((((((.........))))))))))))))).....................((.(((...))).))).)))))))....------ ( -44.70, z-score = -2.74, R) >droSim1.chr3L 4160668 114 - 22553184 GUGAAAUCCGGUGGCUUGUACUUCGGCGGUCGCUUAAUUGGCUUCUAUAACCAAUUGCGGCCGCCACGUCAAAUGUUGUUCUCCUUGGCGCCCUUGGCCCCGGCACCAAAACCU------ (((....((((.((((........(((((((((..((((((.........))))))))))))))).((((((............)))))).....)))))))))))........------ ( -42.00, z-score = -2.68, R) >droSec1.super_2 4611288 114 - 7591821 GUGAAAUCCGGUGGCUUGUACUUCGGCGGUCGCUUAAUUGGCUUCUAUAACCAAUUGCGGCCGCCACGUCAAAUGUUGUUCUCCUUGGCGCCCUUGGCCCCGGCACCGGAACCU------ ......(((((((.(.........(((((((((..((((((.........))))))))))))))).....................((.(((...))).))).)))))))....------ ( -44.70, z-score = -2.74, R) >droYak2.chr3L 5204211 114 - 24197627 GUGAAAUCCGGCGGCUUGUACUUCGGCGGUCGCUUAAUUGGCUUCUAUAACCAAUUGCGGCCGCCACGUCAAAUGUUGUUCUCCUUGGCGCCCUUGGCCCCGGCACCGGAACCU------ ......(((((.(((.........(((((((((..((((((.........)))))))))))))))..(((((............))))))))....((....)).)))))....------ ( -42.10, z-score = -1.68, R) >droEre2.scaffold_4784 7339133 114 - 25762168 GUGAAAUCCGGCGGCUUGUACUUCGGCGGUCGCUUAAUUGGCUUCUAUAACCAAUUGCGGCCGCCACGUCAAAUGUUGUUCUCCUUGGCGCCCUUGGCCCCGGCACCGGAACCC------ ......(((((.(((.........(((((((((..((((((.........)))))))))))))))..(((((............))))))))....((....)).)))))....------ ( -42.10, z-score = -1.60, R) >droAna3.scaffold_13337 2715270 120 - 23293914 GGGAAGUCCGGCGGCUUGUACUUCGGCGGUCGUUUAAUUGGCUUCUAUAACCAAUUGCGGCCGCCACGUCAUAUGUUGUUCUCCUUGGUGCCCUUGGCGCCAGUUCCGGAUCCGGAUCCU ((((.(..(((((...((.((...(((((((((..((((((.........)))))))))))))))..))))..)))))..)))))(((((((...)))))))..((((....)))).... ( -47.90, z-score = -2.49, R) >dp4.chrXR_group8 2741716 114 - 9212921 GUGAAGUCCGGCGGCUUGUACUUCGGCGGUCGCUUAAUUGGCUUCUAUAACCAAUUGCGGCCGCCACGUCAUAUGUUGUUCUCCUUGGCGGCCUUGGCCCCCGAACCGGAGCCG------ ......(((((((((.(((((...(((((((((..((((((.........)))))))))))))))..)).))).))))......((((.(((....))).)))).)))))....------ ( -43.90, z-score = -1.56, R) >droPer1.super_24 1243601 114 + 1556852 GUGAAGUCCGGCGGCUUGUACUUCGGCGGUCGCUUAAUUGGCUUCUAUAACCAAUUGCGGCCGCCACGUCAUAUGUUGUUCUCCUUGGCGGCCUUGGCCCCCGAACCGGAGCCG------ ......(((((((((.(((((...(((((((((..((((((.........)))))))))))))))..)).))).))))......((((.(((....))).)))).)))))....------ ( -43.90, z-score = -1.56, R) >droWil1.scaffold_180698 731153 98 + 11422946 GUAAAAUCUGGCGGCUUGUACUUGGGCGGACGUUUAAUUGGCUUCUAUAACCAAUUGCGGCCGCCACGUCAUAUGUUGUUCUCCUUGGCGCCCUUAGU---------------------- ........((((((((.(((.((((..(((.(((.....))).)))....)))).)))))))))))(((((..............)))))........---------------------- ( -28.44, z-score = -0.60, R) >droVir3.scaffold_12758 241215 113 - 972128 GUAAAGUCCGGCGGCUUGUAUUUGGGCGGUCGCUUAAUUGGCUUCUAUAACCAAUUGCGGCCGCCACGUCAUAUGUUGUUCUCCUUGGCGCCCUUCGACGCCGACGAGGCGCC------- .........((((.(((((.....(((((((((..((((((.........)))))))))))))))....................(((((........)))))))))).))))------- ( -44.60, z-score = -2.06, R) >droMoj3.scaffold_6680 1358051 113 - 24764193 GUAAAAUCCGGCGGCUUGUAUUUGGGCGGUCGCUUAAUUGGCUUCUAUAACCAAUUGCGGCCGCCACGUCAUAUAUUGUUCUCCUUGGCGCCCUUCGAUGCAGCGGCGGCACC------- .......(((.(.(((.(((((.((((((((((..((((((.........))))))))))))((((.(.((.....)).....).))))))))...)))))))).))))....------- ( -40.40, z-score = -1.22, R) >droGri2.scaffold_15110 23409615 113 + 24565398 GUAAAAUCGGGCGGUUUAUAUUUGGGCGGUCGCUUAAUUGGCUUCUAUAACCAAUUGCGGCCGCCACGUCAAAUGUUGUUCUCCUUGGCGCCCCCCUUAGUCGCGGAGGCGCC------- ........(((.((...((((((((((((((((..((((((.........)))))))))))))))....)))))))...)).))).((((((.(((......).)).))))))------- ( -44.80, z-score = -2.34, R) >anoGam1.chrX 12369530 90 + 22145176 -------UUGACGUUGCUUACUACUGCCGUUGCU------GCUCC----AGCACCUGCUGCUCCAACGGCUCCUCCUGCUCCACCCGGCCCGGGUGUACCGGGGGCC------------- -------..................(((((((..------((..(----((...)))..))..)))))))..((((((...((((((...))))))...))))))..------------- ( -33.50, z-score = -0.99, R) >consensus GUGAAAUCCGGCGGCUUGUACUUCGGCGGUCGCUUAAUUGGCUUCUAUAACCAAUUGCGGCCGCCACGUCAAAUGUUGUUCUCCUUGGCGCCCUUGGCCCCGGCACCGGAACC_______ .......((((.((..........(((((((((..((((((.........))))))))))))))).(((((..............))))).))))))....................... (-24.79 = -25.36 + 0.58)
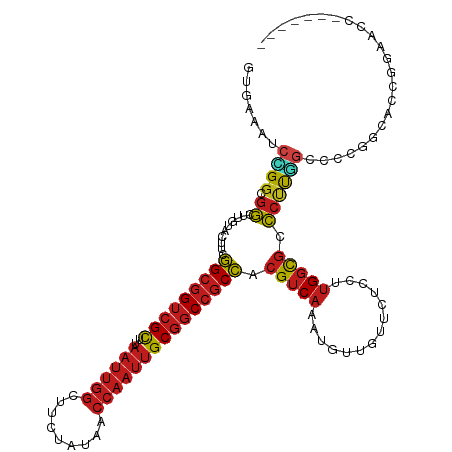
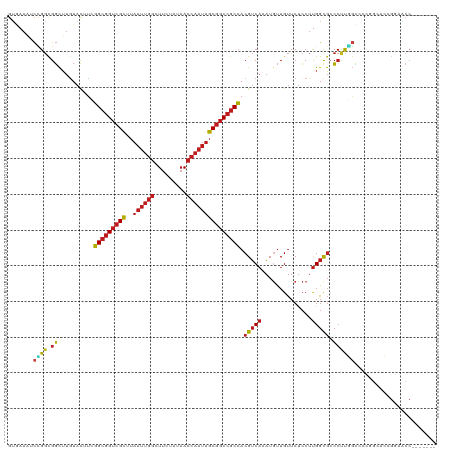
| Location | 4,629,080 – 4,629,192 |
|---|---|
| Length | 112 |
| Sequences | 13 |
| Columns | 123 |
| Reading direction | forward |
| Mean pairwise identity | 81.16 |
| Shannon entropy | 0.42286 |
| G+C content | 0.56686 |
| Mean single sequence MFE | -37.43 |
| Consensus MFE | -21.93 |
| Energy contribution | -22.82 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.778727 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
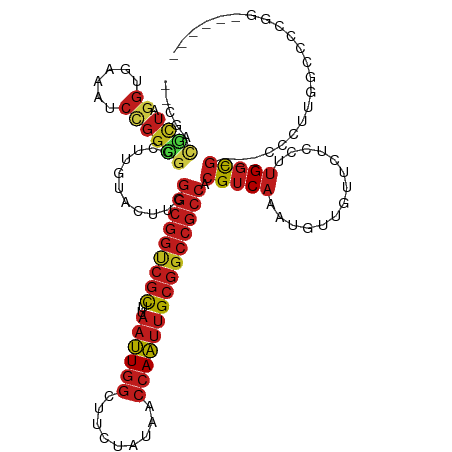
>dm3.chr3L 4629080 112 + 24543557 ------CCGGGGCCAAGGG---CGCCAAGGAGAACAACAUUUGACGUGGCGGCCGCAAUUGGUUAUAGAAGCCAAUUAAGCGACCGCCGAAGUACAAGCCACCGGAUUUCACCUAGUGUCG-- ------(((((((.....(---((.((((..........)))).)))(((((.(((((((((((.....))))))))..))).))))).........))).))))................-- ( -38.30, z-score = -1.65, R) >droSim1.chr3L 4160679 112 + 22553184 ------CCGGGGCCAAGGG---CGCCAAGGAGAACAACAUUUGACGUGGCGGCCGCAAUUGGUUAUAGAAGCCAAUUAAGCGACCGCCGAAGUACAAGCCACCGGAUUUCACCUAGCGUCG-- ------..((.(((...))---).))................((((((((((.(((((((((((.....))))))))..))).)))))(((((.(........).))))).....))))).-- ( -39.70, z-score = -1.96, R) >droSec1.super_2 4611299 112 + 7591821 ------CCGGGGCCAAGGG---CGCCAAGGAGAACAACAUUUGACGUGGCGGCCGCAAUUGGUUAUAGAAGCCAAUUAAGCGACCGCCGAAGUACAAGCCACCGGAUUUCACCUAGCGUCG-- ------..((.(((...))---).))................((((((((((.(((((((((((.....))))))))..))).)))))(((((.(........).))))).....))))).-- ( -39.70, z-score = -1.96, R) >droYak2.chr3L 5204222 112 + 24197627 ------CCGGGGCCAAGGG---CGCCAAGGAGAACAACAUUUGACGUGGCGGCCGCAAUUGGUUAUAGAAGCCAAUUAAGCGACCGCCGAAGUACAAGCCGCCGGAUUUCACCUAGCGUCA-- ------..((.(((...))---).))...............(((((((((((.(((((((((((.....))))))))..))).)))))(((((.(........).))))).....))))))-- ( -39.80, z-score = -1.52, R) >droEre2.scaffold_4784 7339144 112 + 25762168 ------CCGGGGCCAAGGG---CGCCAAGGAGAACAACAUUUGACGUGGCGGCCGCAAUUGGUUAUAGAAGCCAAUUAAGCGACCGCCGAAGUACAAGCCGCCGGAUUUCACCUAGCGUCG-- ------..((.(((...))---).))................((((((((((.(((((((((((.....))))))))..))).)))))(((((.(........).))))).....))))).-- ( -39.70, z-score = -1.43, R) >droAna3.scaffold_13337 2715281 118 + 23293914 CCGGAACUGGCGCCAAGGG---CACCAAGGAGAACAACAUAUGACGUGGCGGCCGCAAUUGGUUAUAGAAGCCAAUUAAACGACCGCCGAAGUACAAGCCGCCGGACUUCCCCUAGCGGCG-- ((.....(((.(((...))---).))).))...........((((.((((((.((.((((((((.....))))))))...)).))))))..)).)).(((((..(.......)..))))).-- ( -38.80, z-score = -0.97, R) >dp4.chrXR_group8 2741727 112 + 9212921 ------CGGGGGCCAAGGC---CGCCAAGGAGAACAACAUAUGACGUGGCGGCCGCAAUUGGUUAUAGAAGCCAAUUAAGCGACCGCCGAAGUACAAGCCGCCGGACUUCACCUAGCACCG-- ------.((.(((....))---).)).(((.................(((((.(((((((((((.....))))))))..))).)))))(((((.(........).))))).))).......-- ( -40.80, z-score = -2.15, R) >droPer1.super_24 1243612 112 - 1556852 ------CGGGGGCCAAGGC---CGCCAAGGAGAACAACAUAUGACGUGGCGGCCGCAAUUGGUUAUAGAAGCCAAUUAAGCGACCGCCGAAGUACAAGCCGCCGGACUUCACCUAGCACCG-- ------.((.(((....))---).)).(((.................(((((.(((((((((((.....))))))))..))).)))))(((((.(........).))))).))).......-- ( -40.80, z-score = -2.15, R) >droWil1.scaffold_180698 731153 109 - 11422946 -----------ACUAAGGG---CGCCAAGGAGAACAACAUAUGACGUGGCGGCCGCAAUUGGUUAUAGAAGCCAAUUAAACGUCCGCCCAAGUACAAGCCGCCAGAUUUUACCUAGCGUCUAA -----------.....(((---(((..(((((((((.....))...(((((((...((((((((.....))))))))....((.(......).))..)))))))..)))).))).)))))).. ( -31.90, z-score = -1.54, R) >droVir3.scaffold_12758 241225 109 + 972128 ------CGGCGUCGAAGGG---CGCCAAGGAGAACAACAUAUGACGUGGCGGCCGCAAUUGGUUAUAGAAGCCAAUUAAGCGACCGCCCAAAUACAAGCCGCCGGACUUUACCUAGCG----- ------.((((((....))---))))...................(.(((((.(((((((((((.....))))))))..))).))))))..........(((..(.......)..)))----- ( -39.10, z-score = -2.43, R) >droMoj3.scaffold_6680 1358061 110 + 24764193 ------CUGCAUCGAAGGG---CGCCAAGGAGAACAAUAUAUGACGUGGCGGCCGCAAUUGGUUAUAGAAGCCAAUUAAGCGACCGCCCAAAUACAAGCCGCCGGAUUUUACCUAGCAU---- ------.(((.......((---((.(..(.....)..........(.(((((.(((((((((((.....))))))))..))).))))))........).))))((......))..))).---- ( -32.20, z-score = -1.14, R) >droGri2.scaffold_15110 23409625 112 - 24565398 ---------CGACUAAGGGGGGCGCCAAGGAGAACAACAUUUGACGUGGCGGCCGCAAUUGGUUAUAGAAGCCAAUUAAGCGACCGCCCAAAUAUAAACCGCCCGAUUUUACCUAAUCCCG-- ---------.......((.(((((....(..(((.....)))..)(.(((((.(((((((((((.....))))))))..))).))))))..........)))))((((......)))))).-- ( -35.10, z-score = -1.91, R) >anoGam1.chrX 12369541 87 - 22145176 ------------CACCCGG----GCCGGGUGGAGCAGGAGGAGCCGUUGGAGCAGCAGGUG-CUGGAGCAGCAACGGCAGUAGUAAGCAACGUCAAUGGCGGCG------------------- ------------((((((.----..))))))..((.......(((((((..(((((....)-))...))..)))))))............(((.....))))).------------------- ( -30.70, z-score = -0.22, R) >consensus ______CCGGGGCCAAGGG___CGCCAAGGAGAACAACAUAUGACGUGGCGGCCGCAAUUGGUUAUAGAAGCCAAUUAAGCGACCGCCGAAGUACAAGCCGCCGGAUUUCACCUAGCGUCG__ ...............(((......((..((....((.....))....(((((.(((((((((((.....))))))))..))).)))))..........))...))......)))......... (-21.93 = -22.82 + 0.89)
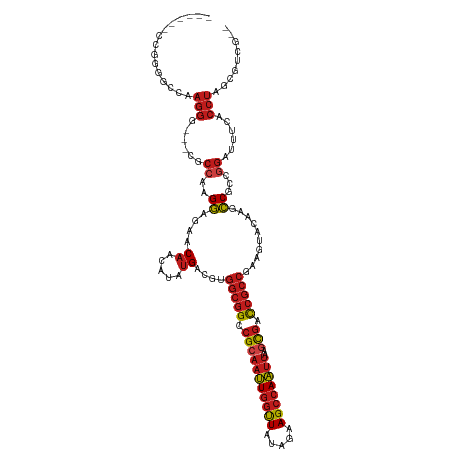
| Location | 4,629,080 – 4,629,192 |
|---|---|
| Length | 112 |
| Sequences | 13 |
| Columns | 123 |
| Reading direction | reverse |
| Mean pairwise identity | 81.16 |
| Shannon entropy | 0.42286 |
| G+C content | 0.56686 |
| Mean single sequence MFE | -39.50 |
| Consensus MFE | -24.93 |
| Energy contribution | -26.18 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.810421 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 4629080 112 - 24543557 --CGACACUAGGUGAAAUCCGGUGGCUUGUACUUCGGCGGUCGCUUAAUUGGCUUCUAUAACCAAUUGCGGCCGCCACGUCAAAUGUUGUUCUCCUUGGCG---CCCUUGGCCCCGG------ --...(((...)))....((((.((((........(((((((((..((((((.........))))))))))))))).((((((............))))))---.....))))))))------ ( -40.80, z-score = -2.05, R) >droSim1.chr3L 4160679 112 - 22553184 --CGACGCUAGGUGAAAUCCGGUGGCUUGUACUUCGGCGGUCGCUUAAUUGGCUUCUAUAACCAAUUGCGGCCGCCACGUCAAAUGUUGUUCUCCUUGGCG---CCCUUGGCCCCGG------ --................((((.((((........(((((((((..((((((.........))))))))))))))).((((((............))))))---.....))))))))------ ( -40.60, z-score = -1.59, R) >droSec1.super_2 4611299 112 - 7591821 --CGACGCUAGGUGAAAUCCGGUGGCUUGUACUUCGGCGGUCGCUUAAUUGGCUUCUAUAACCAAUUGCGGCCGCCACGUCAAAUGUUGUUCUCCUUGGCG---CCCUUGGCCCCGG------ --................((((.((((........(((((((((..((((((.........))))))))))))))).((((((............))))))---.....))))))))------ ( -40.60, z-score = -1.59, R) >droYak2.chr3L 5204222 112 - 24197627 --UGACGCUAGGUGAAAUCCGGCGGCUUGUACUUCGGCGGUCGCUUAAUUGGCUUCUAUAACCAAUUGCGGCCGCCACGUCAAAUGUUGUUCUCCUUGGCG---CCCUUGGCCCCGG------ --................((((.((((........(((((((((..((((((.........))))))))))))))).((((((............))))))---.....))))))))------ ( -40.60, z-score = -1.40, R) >droEre2.scaffold_4784 7339144 112 - 25762168 --CGACGCUAGGUGAAAUCCGGCGGCUUGUACUUCGGCGGUCGCUUAAUUGGCUUCUAUAACCAAUUGCGGCCGCCACGUCAAAUGUUGUUCUCCUUGGCG---CCCUUGGCCCCGG------ --................((((.((((........(((((((((..((((((.........))))))))))))))).((((((............))))))---.....))))))))------ ( -40.60, z-score = -1.29, R) >droAna3.scaffold_13337 2715281 118 - 23293914 --CGCCGCUAGGGGAAGUCCGGCGGCUUGUACUUCGGCGGUCGUUUAAUUGGCUUCUAUAACCAAUUGCGGCCGCCACGUCAUAUGUUGUUCUCCUUGGUG---CCCUUGGCGCCAGUUCCGG --.((((((.(((....))))))))).........(((((((((..((((((.........)))))))))))))))...................((((((---((...))))))))...... ( -46.80, z-score = -1.84, R) >dp4.chrXR_group8 2741727 112 - 9212921 --CGGUGCUAGGUGAAGUCCGGCGGCUUGUACUUCGGCGGUCGCUUAAUUGGCUUCUAUAACCAAUUGCGGCCGCCACGUCAUAUGUUGUUCUCCUUGGCG---GCCUUGGCCCCCG------ --(((.((((((.((.(..(((((...((.((...(((((((((..((((((.........)))))))))))))))..))))..)))))..)))))))))(---((....))).)))------ ( -45.30, z-score = -2.13, R) >droPer1.super_24 1243612 112 + 1556852 --CGGUGCUAGGUGAAGUCCGGCGGCUUGUACUUCGGCGGUCGCUUAAUUGGCUUCUAUAACCAAUUGCGGCCGCCACGUCAUAUGUUGUUCUCCUUGGCG---GCCUUGGCCCCCG------ --(((.((((((.((.(..(((((...((.((...(((((((((..((((((.........)))))))))))))))..))))..)))))..)))))))))(---((....))).)))------ ( -45.30, z-score = -2.13, R) >droWil1.scaffold_180698 731153 109 + 11422946 UUAGACGCUAGGUAAAAUCUGGCGGCUUGUACUUGGGCGGACGUUUAAUUGGCUUCUAUAACCAAUUGCGGCCGCCACGUCAUAUGUUGUUCUCCUUGGCG---CCCUUAGU----------- ......((((((.......((((((((.(((.((((..(((.(((.....))).)))....)))).)))))))))))(((((..............)))))---..))))))----------- ( -33.14, z-score = -0.58, R) >droVir3.scaffold_12758 241225 109 - 972128 -----CGCUAGGUAAAGUCCGGCGGCUUGUAUUUGGGCGGUCGCUUAAUUGGCUUCUAUAACCAAUUGCGGCCGCCACGUCAUAUGUUGUUCUCCUUGGCG---CCCUUCGACGCCG------ -----.(((.((......)))))((((((.....((((((((((..((((((.........))))))))))))((((.(.((.....)).....).)))))---)))..))).))).------ ( -38.10, z-score = -1.20, R) >droMoj3.scaffold_6680 1358061 110 - 24764193 ----AUGCUAGGUAAAAUCCGGCGGCUUGUAUUUGGGCGGUCGCUUAAUUGGCUUCUAUAACCAAUUGCGGCCGCCACGUCAUAUAUUGUUCUCCUUGGCG---CCCUUCGAUGCAG------ ----.((((.((......))))))...((((((.((((((((((..((((((.........))))))))))))((((.(.((.....)).....).)))))---)))...)))))).------ ( -36.50, z-score = -1.41, R) >droGri2.scaffold_15110 23409625 112 + 24565398 --CGGGAUUAGGUAAAAUCGGGCGGUUUAUAUUUGGGCGGUCGCUUAAUUGGCUUCUAUAACCAAUUGCGGCCGCCACGUCAAAUGUUGUUCUCCUUGGCGCCCCCCUUAGUCG--------- --...(((((((.......(((.(((.........(((((((((..((((((.........)))))))))))))))..(((((............)))))))))))))))))).--------- ( -39.11, z-score = -1.65, R) >anoGam1.chrX 12369541 87 + 22145176 -------------------CGCCGCCAUUGACGUUGCUUACUACUGCCGUUGCUGCUCCAG-CACCUGCUGCUCCAACGGCUCCUCCUGCUCCACCCGGC----CCGGGUG------------ -------------------................((........(((((((..((..(((-...)))..))..))))))).......))..((((((..----.))))))------------ ( -26.06, z-score = -0.97, R) >consensus __CGACGCUAGGUGAAAUCCGGCGGCUUGUACUUCGGCGGUCGCUUAAUUGGCUUCUAUAACCAAUUGCGGCCGCCACGUCAAAUGUUGUUCUCCUUGGCG___CCCUUGGCCCCGG______ ......((((((.......................(((((((((..((((((.........))))))))))))))).(((((..............))))).....))))))........... (-24.93 = -26.18 + 1.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:03:23 2011