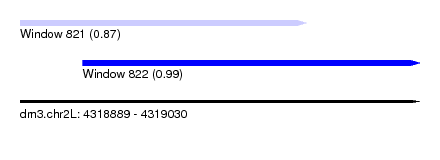
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 4,318,889 – 4,319,030 |
| Length | 141 |
| Max. P | 0.991022 |

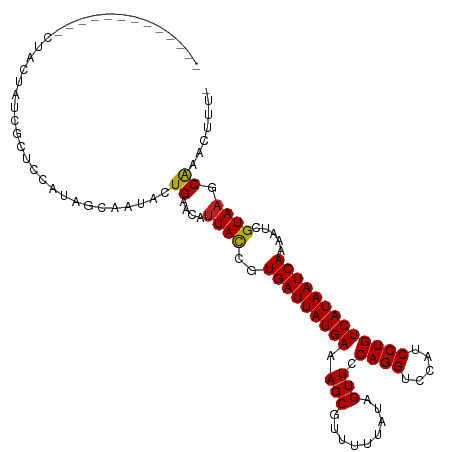
| Location | 4,318,889 – 4,318,990 |
|---|---|
| Length | 101 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 85.48 |
| Shannon entropy | 0.31115 |
| G+C content | 0.35742 |
| Mean single sequence MFE | -17.21 |
| Consensus MFE | -13.86 |
| Energy contribution | -13.68 |
| Covariance contribution | -0.18 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.867316 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 4318889 101 + 23011544 -------------CUACUAUCGCUCCAUAGCAAUACUGAACAUUACCGUGAUUAUGAAAGCGUUUUUAUAGUUCCAGGUCCAUCCUGUCAUAAUCAAAAUCGUAAGCAAACUUU- -------------........(((....))).....((....((((..(((((((((.(((.........))).((((.....))))))))))))).....)))).))......- ( -16.10, z-score = -1.35, R) >droSim1.chr2L 4241065 101 + 22036055 -------------CUACUAUCGCUCCAUAGCAAUACUGAACAUUACCGUGAUUAUGAAAGCGUUUUUAUAGUUCCAGGUCCAUCCUGUCAUAAUCAAAAUCGUAAGCAAACUUU- -------------........(((....))).....((....((((..(((((((((.(((.........))).((((.....))))))))))))).....)))).))......- ( -16.10, z-score = -1.35, R) >droSec1.super_5 2416487 101 + 5866729 -------------CUACUAUUGCUCCAUAGCAAUACUGAACAUUACCGUGAUUAUGAAAGCGUUUUUAUAGUUCCAGGUCCAUCCUGUCAUAAUCAAAAUCGUAAGCAAACUUU- -------------....(((((((....))))))).((....((((..(((((((((.(((.........))).((((.....))))))))))))).....)))).))......- ( -19.60, z-score = -2.45, R) >droYak2.chr2L 4350057 101 + 22324452 -------------CUACUAUCGCUCCAUAGCAAUACUGAACAUUACCGUGAUUAUGAAAGCGUUUUUAUAGUUCCAGGUCCAUCCUGUCAUAAUCAAAAUCGUAAGCAAACUUU- -------------........(((....))).....((....((((..(((((((((.(((.........))).((((.....))))))))))))).....)))).))......- ( -16.10, z-score = -1.35, R) >droEre2.scaffold_4929 4395659 101 + 26641161 -------------CUACUAUCGCUCCAUAGCAAUACUGAACAUUACCGUGAUUAUGAAAGCGUUUUUAUAGUUCCAGGUCCAUCCUGUCAUAAUCAAAAUCGUAAGCAAACUUU- -------------........(((....))).....((....((((..(((((((((.(((.........))).((((.....))))))))))))).....)))).))......- ( -16.10, z-score = -1.35, R) >droAna3.scaffold_12916 1515004 103 + 16180835 -----------UUCUCCACGUCCUCUAUAGCAAUAUUGAACAUUACCGUGAUUAUGAAAGCGUUUUUAUAGUUCCAGGUCCAUCCUGUCAUAAUCAAAAUCGUAAGCGAAUUUU- -----------.......(((..((.(((....))).))...((((..(((((((((.(((.........))).((((.....))))))))))))).....)))))))......- ( -17.00, z-score = -1.62, R) >dp4.chr4_group2 314445 103 + 1235136 -----------CCAUCGCUCUCUCUUAUAGCAAUACUGAACAUUACCGUGAUUAUGAAAGCGUUUUUAUAGUUCCAGGUCCAUCCUGUCAUAAUCAAAAUCGUAAGCAAACAUU- -----------.....(((..(.....(((.....)))..........(((((((((.(((.........))).((((.....))))))))))))).....)..))).......- ( -15.80, z-score = -1.37, R) >droPer1.super_8 391109 103 - 3966273 -----------CCAUCGCUCUCUCUUAUAGCAAUACUGAACAUUACCGUGAUUAUGAAAGCGUUUUUAUAGUUCCAGGUCCAUCCUGUCAUAAUCAAAAUCGUAAGCAAACAUU- -----------.....(((..(.....(((.....)))..........(((((((((.(((.........))).((((.....))))))))))))).....)..))).......- ( -15.80, z-score = -1.37, R) >droWil1.scaffold_180772 2042338 96 + 8906247 ------------------UUAUCUAUAUAGCAAUACUGAACAUUACCGUGAUUAUGAAAGCGUUUUUAUAGUUCCAGGUCCAUCCUGUCAUAAUCAAAAUCGUAAGCAAAACUA- ------------------..................((....((((..(((((((((.(((.........))).((((.....))))))))))))).....)))).))......- ( -14.20, z-score = -0.98, R) >droVir3.scaffold_13246 388453 109 + 2672878 ------ACUACUAUUGCUCGUUAAAAAUAGCAAUACUGAACAUUAUCGUGAUUAUGAAAGCGUUUUUAUAGUUCCAGGUCCAUCCUGUCAUAAUCAAAAUCGUAAGCAAUGAUUU ------.....(((((((.((((....)))).................(((((((((.(((.........))).((((.....)))))))))))))........))))))).... ( -20.30, z-score = -1.77, R) >droMoj3.scaffold_6500 389213 112 + 32352404 CCUUCCACUGCUCUUGC--GCUACCAAUAGCAAUACUGAACACUAUCGUGAUUAUGAAAGCGUUUUUAUAGUUCCAGGUCCAUCCUGUCAUAAUCAAAAUCGUAAGCAAUGUUU- .....((.((((..(((--((((....)))).................(((((((((.(((.........))).((((.....))))))))))))).....))))))).))...- ( -22.20, z-score = -1.99, R) >droGri2.scaffold_15252 337404 99 - 17193109 ---------GCUACUA------AUUAUUAGCAAUACUGAACAUUAUCGUGAUUAUGAAAGCGUUUUUAUAGUUCCAGGUCCAUCCUGUCAUAAUCAAAAUCGUAAGCAAACUCU- ---------(((((..------....((((.....)))).........(((((((((.(((.........))).((((.....))))))))))))).....)).))).......- ( -17.20, z-score = -1.81, R) >consensus _____________CUACUAUCGCUCCAUAGCAAUACUGAACAUUACCGUGAUUAUGAAAGCGUUUUUAUAGUUCCAGGUCCAUCCUGUCAUAAUCAAAAUCGUAAGCAAACUUU_ ....................................((....((((..(((((((((.(((.........))).((((.....))))))))))))).....)))).))....... (-13.86 = -13.68 + -0.18)

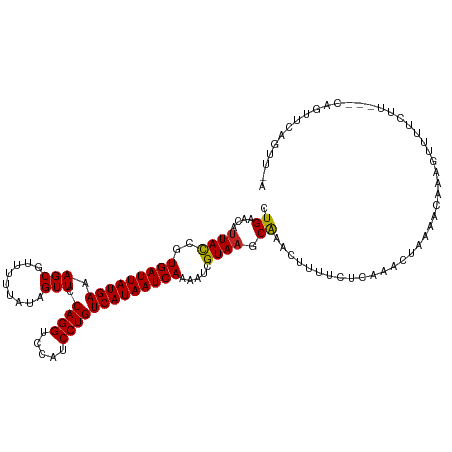
| Location | 4,318,911 – 4,319,030 |
|---|---|
| Length | 119 |
| Sequences | 11 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 83.35 |
| Shannon entropy | 0.36650 |
| G+C content | 0.32992 |
| Mean single sequence MFE | -20.65 |
| Consensus MFE | -13.83 |
| Energy contribution | -13.64 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.45 |
| SVM RNA-class probability | 0.991022 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 4318911 119 + 23011544 CUGAACAUUACCGUGAUUAUGAAAGCGUUUUUAUAGUUCCAGGUCCAUCCUGUCAUAAUCAAAAUCGUAAGCAAACUUUUCUCAAACUUAAACAAAGUUUUCUUCCUCAGUUCAAUUAA .(((((.......(((((((((.(((.........))).((((.....))))))))))))).....(.(((.(((((((..............))))))).))).)...)))))..... ( -21.64, z-score = -3.43, R) >droSim1.chr2L 4241087 119 + 22036055 CUGAACAUUACCGUGAUUAUGAAAGCGUUUUUAUAGUUCCAGGUCCAUCCUGUCAUAAUCAAAAUCGUAAGCAAACUUUUCUCAAACUUAAACAAAGUUUUCUUCUUCAGUUCAGUUAA ((((((.......(((((((((.(((.........))).((((.....))))))))))))).....(.(((.(((((((..............))))))).))))....)))))).... ( -23.64, z-score = -3.76, R) >droSec1.super_5 2416509 119 + 5866729 CUGAACAUUACCGUGAUUAUGAAAGCGUUUUUAUAGUUCCAGGUCCAUCCUGUCAUAAUCAAAAUCGUAAGCAAACUUUUCUCAAACUAAAACAAAGUUUUCUUCUUCAGUUCAGUUAA ((((((.......(((((((((.(((.........))).((((.....))))))))))))).....(.(((.(((((((..............))))))).))))....)))))).... ( -23.64, z-score = -3.80, R) >droYak2.chr2L 4350079 116 + 22324452 CUGAACAUUACCGUGAUUAUGAAAGCGUUUUUAUAGUUCCAGGUCCAUCCUGUCAUAAUCAAAAUCGUAAGCAAACUUUUCUCAAACUUAAACAAAGUUUUCUU---CAGUUCAGUUGA ((((((.......(((((((((.(((.........))).((((.....))))))))))))).....(.(((.(((((((..............))))))).)))---).)))))).... ( -24.14, z-score = -3.17, R) >droEre2.scaffold_4929 4395681 115 + 26641161 CUGAACAUUACCGUGAUUAUGAAAGCGUUUUUAUAGUUCCAGGUCCAUCCUGUCAUAAUCAAAAUCGUAAGCAAACUUUUCUCAAACUUAAACAA-GUUUUCUU---CCGUUCAGUUGA ((((((.......(((((((((.(((.........))).((((.....))))))))))))).....(.(((.((((((...............))-)))).)))---.))))))).... ( -23.56, z-score = -3.06, R) >droAna3.scaffold_12916 1515028 115 + 16180835 UUGAACAUUACCGUGAUUAUGAAAGCGUUUUUAUAGUUCCAGGUCCAUCCUGUCAUAAUCAAAAUCGUAAGCGAAUUUUUCUAAAACUUAAAGGAAGUUUUUUAAUUUGAAGUAG---- .......((((..(((((((((.(((.........))).((((.....))))))))))))).....)))).((((((.....(((((((.....)))))))..))))))......---- ( -20.20, z-score = -1.16, R) >dp4.chr4_group2 314469 99 + 1235136 CUGAACAUUACCGUGAUUAUGAAAGCGUUUUUAUAGUUCCAGGUCCAUCCUGUCAUAAUCAAAAUCGUAAGCAAACAUUUCUCUAACUAAAA----GUUCUCU---------------- ..((((.......(((((((((.(((.........))).((((.....))))))))))))).....((......))................----))))...---------------- ( -16.20, z-score = -2.07, R) >droPer1.super_8 391133 99 - 3966273 CUGAACAUUACCGUGAUUAUGAAAGCGUUUUUAUAGUUCCAGGUCCAUCCUGUCAUAAUCAAAAUCGUAAGCAAACAUUUCUCUAACUAAAA----GUUCUCU---------------- ..((((.......(((((((((.(((.........))).((((.....))))))))))))).....((......))................----))))...---------------- ( -16.20, z-score = -2.07, R) >droVir3.scaffold_13246 388482 118 + 2672878 CUGAACAUUAUCGUGAUUAUGAAAGCGUUUUUAUAGUUCCAGGUCCAUCCUGUCAUAAUCAAAAUCGUAAGCAAUGAUUUUCUAUACUAAACCCAACUUUUUUCAU-CAGUUCAAUUUG .(((((........((((((((.(((.........))).((((.....))))))))))))((((((((.....)))))))).........................-..)))))..... ( -21.10, z-score = -2.62, R) >droMoj3.scaffold_6500 389246 115 + 32352404 CUGAACACUAUCGUGAUUAUGAAAGCGUUUUUAUAGUUCCAGGUCCAUCCUGUCAUAAUCAAAAUCGUAAGCAAUG-UUUUCUAUACUAAACCAAUAUUUUCUCA---AGUUCAGUUUC ((((((((.((..(((((((((.(((.........))).((((.....)))))))))))))..)).)).......(-(((........)))).............---.)))))).... ( -21.60, z-score = -3.05, R) >droGri2.scaffold_15252 337424 110 - 17193109 CUGAACAUUAUCGUGAUUAUGAAAGCGUUUUUAUAGUUCCAGGUCCAUCCUGUCAUAAUCAAAAUCGUAAGCAAAC-UCUUCUCUACUAAACUA-------UUUAA-UAGUUUCAUUUG .((..........(((((((((.(((.........))).((((.....)))))))))))))..........))...-...........((((((-------(...)-))))))...... ( -15.25, z-score = -1.10, R) >consensus CUGAACAUUACCGUGAUUAUGAAAGCGUUUUUAUAGUUCCAGGUCCAUCCUGUCAUAAUCAAAAUCGUAAGCAAACUUUUCUCAAACUAAAACAAAGUUUUCUU___CAGUUCAGUU_A .((....((((..(((((((((.(((.........))).((((.....))))))))))))).....)))).)).............................................. (-13.83 = -13.64 + -0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:16:06 2011