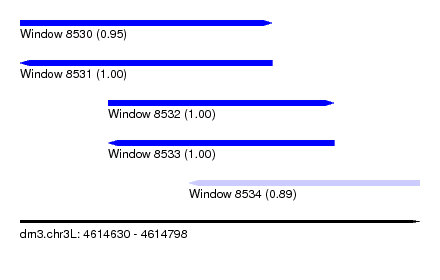
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 4,614,630 – 4,614,798 |
| Length | 168 |
| Max. P | 0.998497 |

| Location | 4,614,630 – 4,614,736 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 79.86 |
| Shannon entropy | 0.34395 |
| G+C content | 0.33263 |
| Mean single sequence MFE | -24.32 |
| Consensus MFE | -13.08 |
| Energy contribution | -13.48 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.947679 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
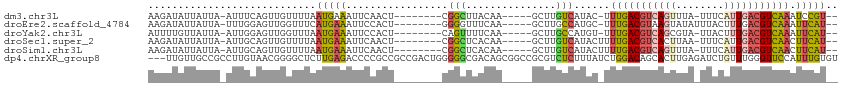
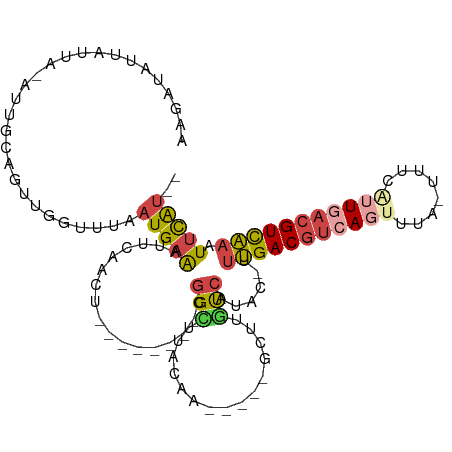
>dm3.chr3L 4614630 106 + 24543557 AACAAAUACGGCGAAUCACCUUAUGAACCUUAUGA---UUAAGGGCACAAAUAAAUUCUUGAA------GACUAUUCGUCACGGAUUUGACGUCAAUG-AAAUAAACUGACGUCAA- .........(((((((...(((.....(((((...---.)))))...(((........)))))------)...)))))))......(((((((((...-........)))))))))- ( -23.80, z-score = -2.66, R) >droEre2.scaffold_4784 7324665 113 + 25762168 AGUACACACGGCGAAUAACUUCAUGACGGUAUUUA--AUUACGGACA-AAAUAAAUUCUGGGAACACAAGGCAAUUCGUCAUGAAUUUGACGUCAAAGUAAAUAUACUUACGUCAA- ...................((((((((((((....--..))).....-..........((....))..........))))))))).(((((((..(((((....))))))))))))- ( -23.30, z-score = -2.07, R) >droYak2.chr3L 5189613 110 + 24197627 AAUAAACACGGCGAAUAACUUUAUGAAGGUAUUUA--CUUACGAGCA-AAAUAAAUUCUCAGAACA--AGGCUAUUCGUCAUGAAUUUGACGUCAAAGUAA-UACGCUGACGUCAA- ......((.((((((((.((((......(((....--..)))(((.(-(......))))).....)--))).)))))))).))...(((((((((..(...-..)..)))))))))- ( -26.70, z-score = -3.18, R) >droSec1.super_2 4596310 108 + 7591821 AAUAAACACGGAGAAUUACUUUAUAAAGUUAUUUA--CUUAAGGGCAUAAAUAAAUUCUUGAA------GACUAUUCGUCAUGAAGUUGACGUCAAUG-AAAUUAAGUGACGUCAAA ......((.(..((((..(((((..((.(((((((--((....))..))))))).))..))))------)...))))..).))...(((((((((.(.-......).))))))))). ( -20.50, z-score = -1.61, R) >droSim1.chr3L 4146108 110 + 22553184 AAUAAACACGGCGAAUAACUUUAUGAAGGUAUUUAUACUUUAGGGCACAAAUAAAUUCUUGAA------GACAAUUCGUCAUGAAGUUGACGUCAAUG-AAAUAAACUGACGUCAAA ......((.(((((((..(((((.(((..(((((...((....))...)))))..))).))))------)...))))))).))...(((((((((...-........))))))))). ( -27.30, z-score = -3.80, R) >consensus AAUAAACACGGCGAAUAACUUUAUGAAGGUAUUUA__CUUAAGGGCA_AAAUAAAUUCUUGAA______GACUAUUCGUCAUGAAUUUGACGUCAAUG_AAAUAAACUGACGUCAA_ .........(((((((.........................................................)))))))......(((((((((............))))))))). (-13.08 = -13.48 + 0.40)

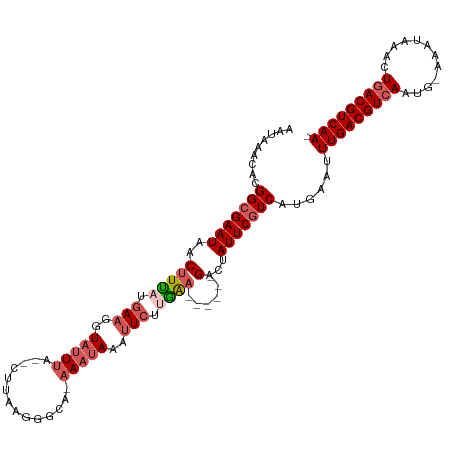
| Location | 4,614,630 – 4,614,736 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 79.86 |
| Shannon entropy | 0.34395 |
| G+C content | 0.33263 |
| Mean single sequence MFE | -26.42 |
| Consensus MFE | -14.28 |
| Energy contribution | -15.16 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.25 |
| Mean z-score | -3.52 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.89 |
| SVM RNA-class probability | 0.996173 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 4614630 106 - 24543557 -UUGACGUCAGUUUAUUU-CAUUGACGUCAAAUCCGUGACGAAUAGUC------UUCAAGAAUUUAUUUGUGCCCUUAA---UCAUAAGGUUCAUAAGGUGAUUCGCCGUAUUUGUU -(((((((((((......-.)))))))))))......((((((((...------.............(((((.(((((.---...)))))..)))))((((...)))).)))))))) ( -27.70, z-score = -3.25, R) >droEre2.scaffold_4784 7324665 113 - 25762168 -UUGACGUAAGUAUAUUUACUUUGACGUCAAAUUCAUGACGAAUUGCCUUGUGUUCCCAGAAUUUAUUU-UGUCCGUAAU--UAAAUACCGUCAUGAAGUUAUUCGCCGUGUGUACU -((((((((((((....)))))..))))))).(((((((((................(((((....)))-))...(((..--....))))))))))))................... ( -23.70, z-score = -2.02, R) >droYak2.chr3L 5189613 110 - 24197627 -UUGACGUCAGCGUA-UUACUUUGACGUCAAAUUCAUGACGAAUAGCCU--UGUUCUGAGAAUUUAUUU-UGCUCGUAAG--UAAAUACCUUCAUAAAGUUAUUCGCCGUGUUUAUU -((((((((((.(..-...).))))))))))...((((.((((((((.(--(((...(.(.((((((((-.......)))--))))).))...)))).)))))))).))))...... ( -29.80, z-score = -3.96, R) >droSec1.super_2 4596310 108 - 7591821 UUUGACGUCACUUAAUUU-CAUUGACGUCAACUUCAUGACGAAUAGUC------UUCAAGAAUUUAUUUAUGCCCUUAAG--UAAAUAACUUUAUAAAGUAAUUCUCCGUGUUUAUU .(((((((((........-...)))))))))...((((..((((....------.......(((((((((......))))--))))).((((....)))).))))..))))...... ( -20.30, z-score = -2.67, R) >droSim1.chr3L 4146108 110 - 22553184 UUUGACGUCAGUUUAUUU-CAUUGACGUCAACUUCAUGACGAAUUGUC------UUCAAGAAUUUAUUUGUGCCCUAAAGUAUAAAUACCUUCAUAAAGUUAUUCGCCGUGUUUAUU .(((((((((((......-.)))))))))))...((((.(((((...(------((...(((..(((((((((......)))))))))..)))...)))..))))).))))...... ( -30.60, z-score = -5.67, R) >consensus _UUGACGUCAGUUUAUUU_CAUUGACGUCAAAUUCAUGACGAAUAGUC______UUCAAGAAUUUAUUU_UGCCCUUAAG__UAAAUACCUUCAUAAAGUUAUUCGCCGUGUUUAUU .(((((((((((........)))))))))))............................((((...((((((...(((........)))...))))))...))))............ (-14.28 = -15.16 + 0.88)

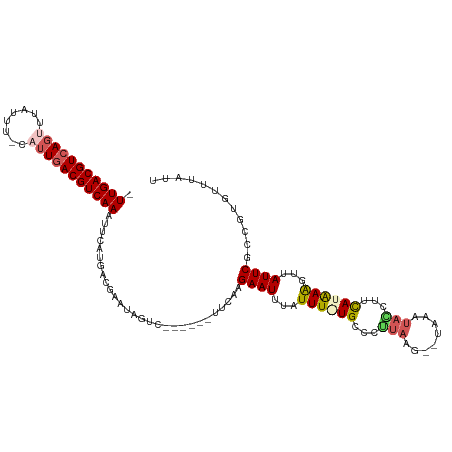
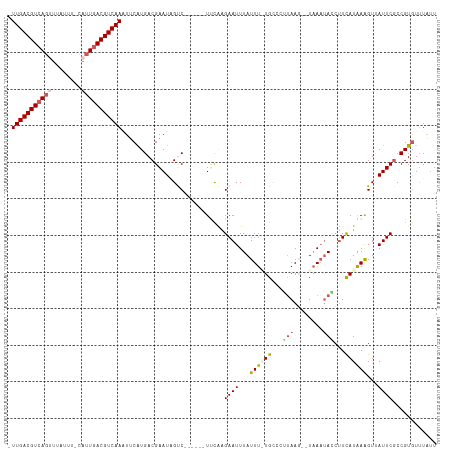
| Location | 4,614,667 – 4,614,762 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 66.43 |
| Shannon entropy | 0.57163 |
| G+C content | 0.40982 |
| Mean single sequence MFE | -26.77 |
| Consensus MFE | -13.15 |
| Energy contribution | -14.20 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.38 |
| SVM RNA-class probability | 0.998497 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 4614667 95 + 24543557 AAGGGCACAAAUAAAUUCUUGAA------GACUAUUCGUCACG-GAUUUGACGUCAAUGAAA--UAAACUGACGUCAAA-GUAUGACAAGC-----UUGU-AAGCCGAGUU------- ...(((((((..........(((------.....)))((((..-..((((((((((......--.....))))))))))-...))))....-----))))-..))).....------- ( -23.80, z-score = -2.23, R) >droEre2.scaffold_4784 7324703 101 + 25762168 ACGGACA-AAAUAAAUUCUGGGAACACAAGGCAAUUCGUCAUG-AAUUUGACGUCAAAGUAAA-UAUACUUACGUCAAA-GCAUGGCAAGC-----UUGA-AACCCCAGUG------- .......-.........(((((......((((.....((((((-..((((((((..(((((..-..)))))))))))))-.))))))..))-----))..-...)))))..------- ( -28.50, z-score = -3.79, R) >droYak2.chr3L 5189651 98 + 24197627 ACGAGCA-AAAUAAAUUCUCAGAACA--AGGCUAUUCGUCAUG-AAUUUGACGUCAAAGUAA--UACGCUGACGUCAAA-ACAUGGCAAGC-----UUGA-AAACUGAGUG------- .......-.........(((((....--(((((....((((((-..((((((((((..(...--..)..))))))))))-.)))))).)))-----))..-...)))))..------- ( -30.20, z-score = -4.21, R) >droSec1.super_2 4596348 96 + 7591821 AAGGGCAUAAAUAAAUUCUUGAA------GACUAUUCGUCAUG-AAGUUGACGUCAAUGAAA--UUAAGUGACGUCAAAAGUAUGACAAGC-----UUGU-GAGCCGAGUU------- ...(((..............(((------.....)))((((((-...(((((((((.(....--...).)))))))))...))))))....-----....-..))).....------- ( -26.30, z-score = -3.08, R) >droSim1.chr3L 4146148 96 + 22553184 UAGGGCACAAAUAAAUUCUUGAA------GACAAUUCGUCAUG-AAGUUGACGUCAAUGAAA--UAAACUGACGUCAAAAGUAUGACAAGC-----UUGU-GAGCCGAGUU------- ..((.(((((..........(((------.....)))((((((-...(((((((((......--.....)))))))))...))))))....-----))))-)..)).....------- ( -28.20, z-score = -3.65, R) >dp4.chrXR_group8 2725797 106 + 9212921 --------AUAUCAUUUCCAGAGC----AAACACUCCGCCACACAAAUGGAACCCAAACAGAUCUCAAGUGCUGUCCAGAUAAAGAGACGCGGCCGCUGUCGCCCCCAGUCGGCGGCG --------............(((.----.....)))(((........(((...)))......((((.....((....)).....)))).)))(((((((.(.......).))))))). ( -23.60, z-score = 0.16, R) >consensus AAGGGCA_AAAUAAAUUCUUGAA______GACAAUUCGUCAUG_AAUUUGACGUCAAAGAAA__UAAACUGACGUCAAA_GUAUGACAAGC_____UUGU_AAGCCGAGUU_______ .....................................((((((....(((((((((.............)))))))))...))))))............................... (-13.15 = -14.20 + 1.06)

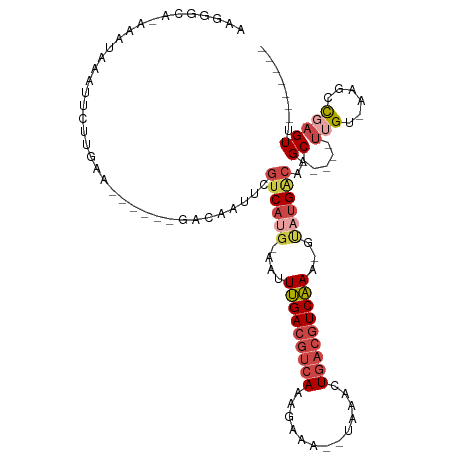
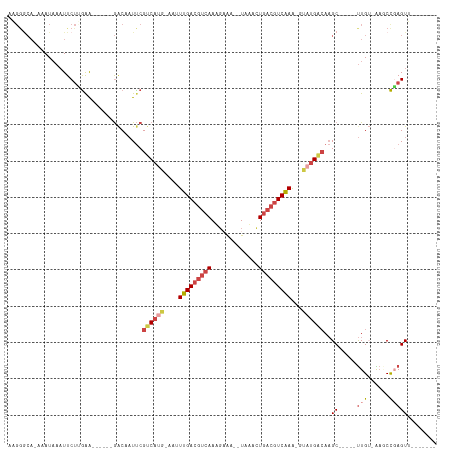
| Location | 4,614,667 – 4,614,762 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 66.43 |
| Shannon entropy | 0.57163 |
| G+C content | 0.40982 |
| Mean single sequence MFE | -27.87 |
| Consensus MFE | -11.27 |
| Energy contribution | -12.52 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.18 |
| Mean z-score | -3.00 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.98 |
| SVM RNA-class probability | 0.996759 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 4614667 95 - 24543557 -------AACUCGGCUU-ACAA-----GCUUGUCAUAC-UUUGACGUCAGUUUA--UUUCAUUGACGUCAAAUC-CGUGACGAAUAGUC------UUCAAGAAUUUAUUUGUGCCCUU -------.....(((..-((((-----(.(((((((..-((((((((((((...--....))))))))))))..-.)))))))....((------.....)).....))))))))... ( -26.30, z-score = -3.84, R) >droEre2.scaffold_4784 7324703 101 - 25762168 -------CACUGGGGUU-UCAA-----GCUUGCCAUGC-UUUGACGUAAGUAUA-UUUACUUUGACGUCAAAUU-CAUGACGAAUUGCCUUGUGUUCCCAGAAUUUAUUU-UGUCCGU -------..((((((..-.(((-----(((((.((((.-(((((((((((((..-..)))))..))))))))..-)))).)))...).))))...)))))).........-....... ( -29.50, z-score = -3.45, R) >droYak2.chr3L 5189651 98 - 24197627 -------CACUCAGUUU-UCAA-----GCUUGCCAUGU-UUUGACGUCAGCGUA--UUACUUUGACGUCAAAUU-CAUGACGAAUAGCCU--UGUUCUGAGAAUUUAUUU-UGCUCGU -------..(((((...-.(((-----(((((.((((.-(((((((((((.(..--...).)))))))))))..-)))).))...)).))--))..))))).........-....... ( -29.00, z-score = -3.66, R) >droSec1.super_2 4596348 96 - 7591821 -------AACUCGGCUC-ACAA-----GCUUGUCAUACUUUUGACGUCACUUAA--UUUCAUUGACGUCAACUU-CAUGACGAAUAGUC------UUCAAGAAUUUAUUUAUGCCCUU -------.....(((..-....-----....(((((....(((((((((.....--......)))))))))...-.)))))(((((.((------.....))...)))))..)))... ( -21.10, z-score = -3.12, R) >droSim1.chr3L 4146148 96 - 22553184 -------AACUCGGCUC-ACAA-----GCUUGUCAUACUUUUGACGUCAGUUUA--UUUCAUUGACGUCAACUU-CAUGACGAAUUGUC------UUCAAGAAUUUAUUUGUGCCCUA -------.....(((.(-((((-----..(((((((....(((((((((((...--....)))))))))))...-.))))))).)))((------.....)).......)).)))... ( -26.50, z-score = -4.00, R) >dp4.chrXR_group8 2725797 106 - 9212921 CGCCGCCGACUGGGGGCGACAGCGGCCGCGUCUCUUUAUCUGGACAGCACUUGAGAUCUGUUUGGGUUCCAUUUGUGUGGCGGAGUGUUU----GCUCUGGAAAUGAUAU-------- ((((.((....)).))))......(((((((((........)))).(((..((..((((....))))..))..))))))))(((((....----)))))...........-------- ( -34.80, z-score = 0.04, R) >consensus _______AACUCGGCUC_ACAA_____GCUUGUCAUAC_UUUGACGUCAGUUUA__UUUCAUUGACGUCAAAUU_CAUGACGAAUAGUC______UUCAAGAAUUUAUUU_UGCCCUU .............................(((((((....((((((((((...........)))))))))).....)))))))................................... (-11.27 = -12.52 + 1.25)


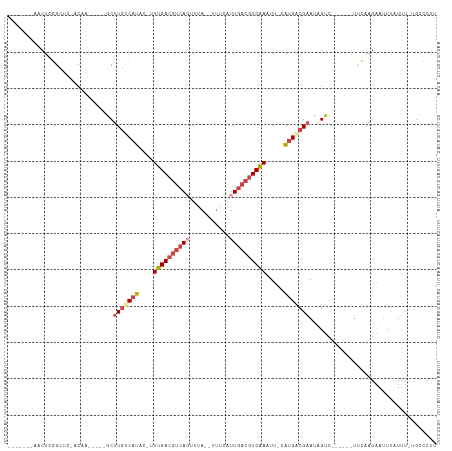
| Location | 4,614,701 – 4,614,798 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 67.67 |
| Shannon entropy | 0.56115 |
| G+C content | 0.36631 |
| Mean single sequence MFE | -25.22 |
| Consensus MFE | -9.20 |
| Energy contribution | -10.45 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.894083 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 4614701 97 - 24543557 AAGAUAUUAUUA-AUUUCAGUUGUUUUAAUGAAAUUCAACU--------CGGCUUACAA-----GCUUGUCAUAC-UUUGACGUCAGUUUA-UUUCAUUGACGUCAAAUCCGU-- ..((((......-.....(((((..((.....))..)))))--------.((((....)-----)))))))....-((((((((((((...-....)))))))))))).....-- ( -21.60, z-score = -3.05, R) >droEre2.scaffold_4784 7324742 98 - 25762168 AAGAUAUUAUUA-UUUGGAGUUGGUUUCAUGAAAUUCCACU--------GGGGUUUCAA-----GCUUGCCAUGC-UUUGACGUAAGUAUAUUUACUUUGACGUCAAAUUCAU-- ............-..((((((((((..(.(((((((((...--------))))))))).-----)...)))).))-(((((((((((((....)))))..)))))))))))).-- ( -25.20, z-score = -2.56, R) >droYak2.chr3L 5189688 97 - 24197627 AUUUUGUUAUUA-AUUGGAGUUGGUUUAAUGAAAUUCCACU--------CAGUUUUCAA-----GCUUGCCAUGU-UUUGACGUCAGCGUA-UUACUUUGACGUCAAAUUCAU-- ..((((..(((.-(.(((((((..........))))))).)--------.)))...)))-----)......(((.-(((((((((((.(..-...).)))))))))))..)))-- ( -20.80, z-score = -1.74, R) >droSec1.super_2 4596382 98 - 7591821 AAGAUAUUAUUA-AUUGCAGUUGUUUUAAUGAAAUUCAACU--------CGGCUCACAA-----GCUUGUCAUACUUUUGACGUCACUUAA-UUUCAUUGACGUCAACUUCAU-- ............-..((.(((((..(((((((((((.....--------.((((....)-----))).((((......)))).......))-)))))))))...))))).)).-- ( -20.30, z-score = -2.60, R) >droSim1.chr3L 4146182 98 - 22553184 AAGAUAUUAUUA-AUUGCAGUUGUUUUAAUGAAAUUCAACU--------CGGCUCACAA-----GCUUGUCAUACUUUUGACGUCAGUUUA-UUUCAUUGACGUCAACUUCAU-- ............-..((.(((((..((((((((((..((((--------.((((....)-----))).((((......))))...)))).)-)))))))))...))))).)).-- ( -21.50, z-score = -2.68, R) >dp4.chrXR_group8 2725825 112 - 9212921 ---UUGUUGCCGCCUUGUAACGGGGCUCUUGAGACCCCGCCGCCGACUGGGGGCGACAGCGGCCGCGUCUCUUUAUCUGGACAGCACUUGAGAUCUGUUUGGGUUCCAUUUGUGU ---..((.(((((..(((...((((((....)).))))(((.((....)).))).)))))))).))((((........)))).((((.((..((((....))))..))...)))) ( -41.90, z-score = -0.67, R) >consensus AAGAUAUUAUUA_AUUGCAGUUGGUUUAAUGAAAUUCAACU________CGGCUUACAA_____GCUUGUCAUAC_UUUGACGUCAGUUUA_UUUCAUUGACGUCAAAUUCAU__ ..................(((((..((.....))..)))))....................................(((((((((((........)))))))))))........ ( -9.20 = -10.45 + 1.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:03:13 2011