
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 4,602,093 – 4,602,150 |
| Length | 57 |
| Max. P | 0.601319 |

| Location | 4,602,093 – 4,602,150 |
|---|---|
| Length | 57 |
| Sequences | 13 |
| Columns | 63 |
| Reading direction | reverse |
| Mean pairwise identity | 79.54 |
| Shannon entropy | 0.47497 |
| G+C content | 0.43367 |
| Mean single sequence MFE | -12.78 |
| Consensus MFE | -6.70 |
| Energy contribution | -7.41 |
| Covariance contribution | 0.71 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.601319 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 4602093 57 - 24543557 CAUUACGGCAUUUACAGUUAAUGCCU---UUAUGUCCUCGAGGAUUUGGC-CUGACAUGCG-- ......((((((.......)))))).---.((((((....(((......)-))))))))..-- ( -13.40, z-score = -1.33, R) >droSim1.chr3L 4133732 57 - 22553184 CAUUACGGCAUUUACAGUUAAUGCCU---UUAUGUCCUCGAGGAUUUGGC-CUGACAUGCG-- ......((((((.......)))))).---.((((((....(((......)-))))))))..-- ( -13.40, z-score = -1.33, R) >droSec1.super_2 4583949 57 - 7591821 CAUUACGGCAUUUACAGUUAAUGCCU---UUAUGUCCUCGAGGAUUUGGC-CUGACAUGCG-- ......((((((.......)))))).---.((((((....(((......)-))))))))..-- ( -13.40, z-score = -1.33, R) >droYak2.chr3L 5176871 57 - 24197627 CAUUACGGCAUUUACAGUUAAUGCCU---UUAUGUCCUCGAGGAUUUGGC-CUGACAUGCG-- ......((((((.......)))))).---.((((((....(((......)-))))))))..-- ( -13.40, z-score = -1.33, R) >droEre2.scaffold_4784 7311883 57 - 25762168 CAUUACGGCAUUUACAGUUAAUGCCU---UUAUGUCCUCGAGGAUUUGGC-CUGACAUGCG-- ......((((((.......)))))).---.((((((....(((......)-))))))))..-- ( -13.40, z-score = -1.33, R) >droAna3.scaffold_13337 2681695 57 - 23293914 CAUUAUGGCAUUUACAGCUAAUGCCU---UUAUGUCCUCAAGGAUUUGGC-CUGACAUGCG-- ......((((((.......)))))).---.((((((....(((......)-))))))))..-- ( -13.10, z-score = -0.92, R) >dp4.chrXR_group8 2711419 57 - 9212921 CAUUACGGCAUUUACAGUUAAUGCCU---UUAUGUCCUCGAGGAUUUGCC-UUGACAUGCG-- ......((((((.......)))))).---.((((((...((((.....))-))))))))..-- ( -14.90, z-score = -2.54, R) >droPer1.super_24 1213574 57 + 1556852 CAUUACGGCAUUUACAGUUAAUGCCU---UUAUGUCCUCGAGGAUUUGCC-UUGACAUGCG-- ......((((((.......)))))).---.((((((...((((.....))-))))))))..-- ( -14.90, z-score = -2.54, R) >droWil1.scaffold_180698 709049 61 + 11422946 UAUAAAGGCAUUUACAGUUUAAGCCUUGAUGAUGUACUUAAGGAUUUGACAAUGACAUGCG-- .......(((((((..(((....((((((........))))))....)))..))).)))).-- ( -11.50, z-score = -1.68, R) >droMoj3.scaffold_6680 1332048 57 - 24764193 CAUUACGGCAUUUACAGUUAAUGCCU---UUAUGUCCUCGAGGAUUGGCUCACGCUGUCA--- .............(((((....(((.---....((((....)))).)))....)))))..--- ( -13.00, z-score = -1.09, R) >droVir3.scaffold_12758 212329 58 - 972128 CAUUACGGCAUUUACAGUUAAUGCCU--GAAAUGUCCUCGAGGAUU-GCCGCACACAGUCA-- .....(((((....(((.......))--)....((((....)))))-))))..........-- ( -11.70, z-score = -0.49, R) >droGri2.scaffold_15110 23382388 55 + 24565398 CAUUACGGCAUUUACAGUUAAUGCCU---UUAUGUCCUCGAGUAUU--GCCUUGCCUUGC--- (((...((((((.......)))))).---..)))....((((....--..))))......--- ( -8.40, z-score = -0.16, R) >apiMel3.Group6 13661853 59 + 14581788 CAUUGCUCCA-UUGCAUUCAAUUCUU---CUGCAAUCAAAAGGAGUAAACGUCAUAUUAUGAA ..((((((((-(((((..........---.)))))).....)))))))............... ( -11.60, z-score = -2.23, R) >consensus CAUUACGGCAUUUACAGUUAAUGCCU___UUAUGUCCUCGAGGAUUUGGC_CUGACAUGCG__ ......((((((.......))))))........((((....)))).................. ( -6.70 = -7.41 + 0.71)

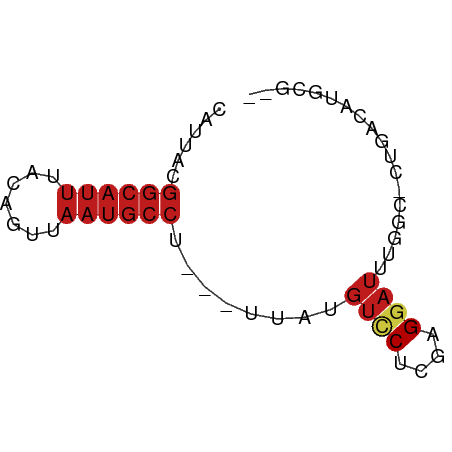
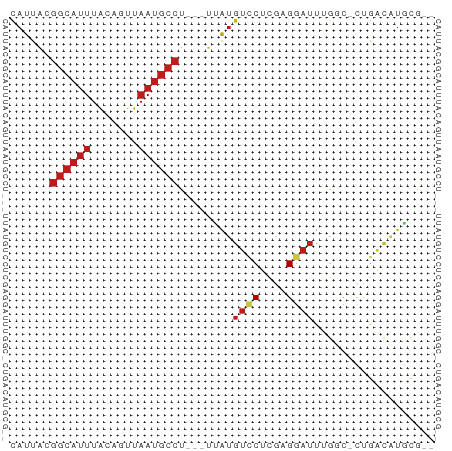
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:03:08 2011