| Sequence ID | dm3.chr3L |
|---|---|
| Location | 4,590,588 – 4,590,671 |
| Length | 83 |
| Max. P | 0.905619 |

| Location | 4,590,588 – 4,590,671 |
|---|---|
| Length | 83 |
| Sequences | 8 |
| Columns | 89 |
| Reading direction | forward |
| Mean pairwise identity | 53.52 |
| Shannon entropy | 0.91543 |
| G+C content | 0.36432 |
| Mean single sequence MFE | -15.45 |
| Consensus MFE | -3.42 |
| Energy contribution | -3.98 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.91 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.22 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.905619 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
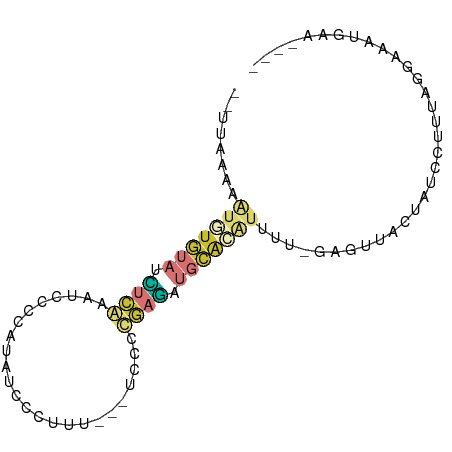
>dm3.chr3L 4590588 83 + 24543557 --UUAAAAAUGUGUAUCUCAAAUCCCCAUAUCCCUUU---CCCCUGAGAUGCACAUUUU-GAGUUACUAUCCUUUAGGAAAUGCUGGAA --...(((((((((((((((.................---....)))))))))))))))-.(((.....(((....)))...))).... ( -20.50, z-score = -3.83, R) >droEre2.scaffold_4784 7300331 79 + 25762168 --UUAAAAAUGUGUAUCUCAAAUUCCCCUAUCCCUCC---CCCC-GAGAUGCACAUUUUAGAGUUAGUUUCCUUUUGGGAAUGAA---- --...((((((((((((((..................---....-)))))))))))))).......((((((....))))))...---- ( -21.65, z-score = -4.69, R) >droYak2.chr3L 5165101 79 + 24197627 --UUAAAAAUAUGUAUCUCAAAUCCCAUAGCCCUCCC---UCCC-GAGAUGCACAUUUUAGAGUUACUUUCCUUUAGGGAAUGAA---- --...(((((.((((((((..................---....-)))))))).)))))........(((((....)))))....---- ( -13.65, z-score = -1.30, R) >droSec1.super_2 4572536 79 + 7591821 --UUAAAAAUGUGUAUCUCAAAUCCCCAUAUCCCUUU---UCCCUGAGCUGCACAUUUU-GAGUUACUAUCCUUUAGGAAAUGAA---- --...((((((((((.((((.................---....)))).))))))))))-.........(((....)))......---- ( -15.30, z-score = -3.09, R) >droSim1.chr3L 4122195 79 + 22553184 --UUAAAAAUGUGUAUCUCAAAUCCCCAUAUCCCUUU---UCCCUGAGCUGCACGUUUU-GAGUUACUUACCUUUAGGAAAUGAA---- --...((((((((((.((((.................---....)))).))))))))))-..........((....)).......---- ( -13.50, z-score = -2.23, R) >droWil1.scaffold_181089 9338961 74 - 12369635 ----------AUAUA-GCCGCUGAUCUGUACCCAUAU---CGAGCUAUAUCUAAAUUCU-GAUCUGUAAUUCUUUCAAAUAUGUUGGAA ----------(((((-((...((((.((....)).))---)).))))))).....(((.-.((.(((...........))).))..))) ( -9.80, z-score = -0.20, R) >droVir3.scaffold_12758 191540 72 + 972128 ---------------AAGUGCGAUUUCAAAUGCAUUG-AUUCCACAAUAUGGAAAAGUCAGGAGUACUAUU-UAUAAAAUAUAAUUCAA ---------------.(((((....((((.....)))-)(((((.....))))).........)))))...-................. ( -9.80, z-score = -0.53, R) >triCas2.ChLG1X 7274052 88 - 8109244 CGACAAAUGUAUGUUUGUUGCUUAUUUUUAGCCGCCACAAUGCGCAACGUGCGUAUUUU-AAGCUCGUCACCUUGGAGGGAUAUUUGCA ((((((((....))))))))..........((.((...(((((((.....)))))))..-..))..(((.((.....)))))....)). ( -19.40, z-score = 0.48, R) >consensus __UUAAAAAUGUGUAUCUCAAAUCCCCAUAUCCCUUU___UCCCCGAGAUGCACAUUUU_GAGUUACUAUCCUUUAGGAAAUGAA____ ........(((((((.((((........................)))).)))))))................................. ( -3.42 = -3.98 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:03:07 2011