
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 4,580,563 – 4,580,656 |
| Length | 93 |
| Max. P | 0.999197 |

| Location | 4,580,563 – 4,580,656 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 88.51 |
| Shannon entropy | 0.20869 |
| G+C content | 0.51215 |
| Mean single sequence MFE | -41.82 |
| Consensus MFE | -38.01 |
| Energy contribution | -38.82 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.13 |
| Mean z-score | -4.61 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.70 |
| SVM RNA-class probability | 0.999197 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
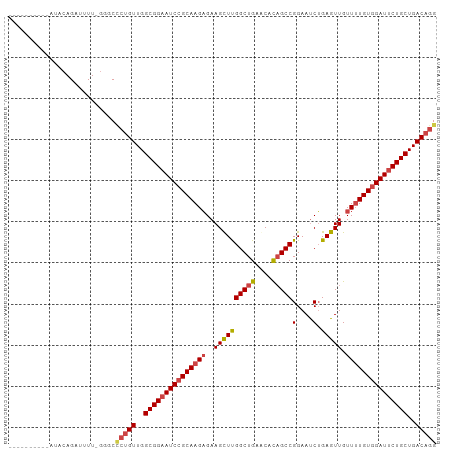
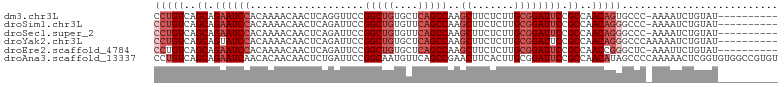
>dm3.chr3L 4580563 93 + 24543557 ----------AUACAGAUUUU-GGGCACUGUUGGCGGAAUCCGCAAGAGAAGCUUGGCUGAGCACAGCCGGAACCUGAGUUGUUUUGUGGAUUCUGCUGACAGG ----------...........-.....((((..(((((((((((((((..((((((((((....)))))(....).))))).)))))))))))))))..)))). ( -44.40, z-score = -5.63, R) >droSim1.chr3L 4112212 93 + 22553184 ----------AUACAGAUUUU-GGGCCCUGUUGGCGGAAUCCGCAAGAGAAGCUUGGCUGAACACAGCCGGAAUCUGAGUUGUUUUGUGGAUUCUGCUGACAGG ----------...........-....(((((..(((((((((((((((..((((((((((....))))).......))))).)))))))))))))))..))))) ( -44.81, z-score = -5.91, R) >droSec1.super_2 4562593 93 + 7591821 ----------AUACAGAUUUU-GGGCCCUGUUGGCGGAAUCCGCAAGAGAAGCUUGGCUGAACACAGCCGGAAUCUGAGUUGUUUUGUGGAUUCUGCUGACAGG ----------...........-....(((((..(((((((((((((((..((((((((((....))))).......))))).)))))))))))))))..))))) ( -44.81, z-score = -5.91, R) >droYak2.chr3L 5154739 94 + 24197627 ----------AUACAGAUUUUUGGGCCCUGUUGGCGGAAUCCGCAAGAGAAGCUUGGCUGAGCACAGCCGGAAUCUGAGUUGUUUUGUGGAUACUGCUGACAGG ----------................(((((..((((.((((((((((..((((((((((....))))).......))))).)))))))))).))))..))))) ( -40.41, z-score = -4.36, R) >droEre2.scaffold_4784 7290366 93 + 25762168 ----------AUACAGAAUUU-GAGCCCGGUUGGCGGAAUCCGCAAGAGAAGCUUGGCUGAGCACAGCCGGAAUCUGAGUUGUUUUGUGGAUUCUGCUGACAGG ----------...........-....((.((..(((((((((((((((..((((((((((....))))).......))))).)))))))))))))))..)).)) ( -40.21, z-score = -4.29, R) >droAna3.scaffold_13337 2658497 104 + 23293914 ACACGGCCACACCGAGUUUUUGGGGCUAUGUUGGCGGAAUCCGCAAGUGAAGUUCGGCUGAACAUUGCCGGAAUCAGAGUUGUUGUGUUGAUUCUGCUGACAGG ....((((...((((....)))))))).(((..((((((((.(((((..(..((((((........)))))).......)..)).))).))))))))..))).. ( -36.30, z-score = -1.53, R) >consensus __________AUACAGAUUUU_GGGCCCUGUUGGCGGAAUCCGCAAGAGAAGCUUGGCUGAACACAGCCGGAAUCUGAGUUGUUUUGUGGAUUCUGCUGACAGG ..........................(((((..(((((((((((((((..((((((((((....)))))(....).))))).)))))))))))))))..))))) (-38.01 = -38.82 + 0.81)


| Location | 4,580,563 – 4,580,656 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 88.51 |
| Shannon entropy | 0.20869 |
| G+C content | 0.51215 |
| Mean single sequence MFE | -27.00 |
| Consensus MFE | -22.52 |
| Energy contribution | -24.02 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.959752 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
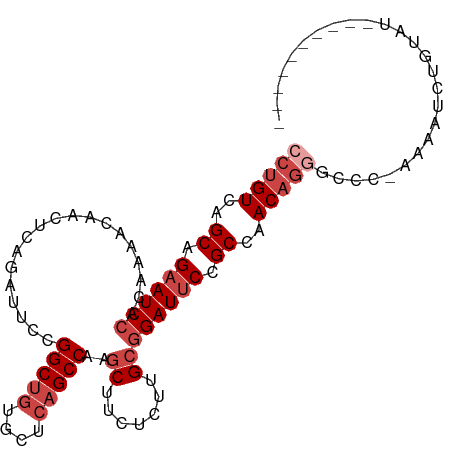
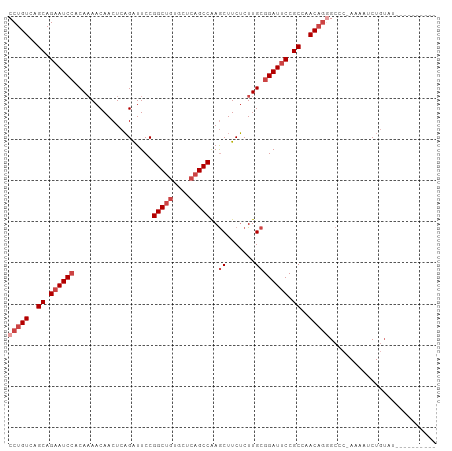
>dm3.chr3L 4580563 93 - 24543557 CCUGUCAGCAGAAUCCACAAAACAACUCAGGUUCCGGCUGUGCUCAGCCAAGCUUCUCUUGCGGAUUCCGCCAACAGUGCCC-AAAAUCUGUAU---------- .((((..((.((((((...................(((((....)))))..((.......)))))))).))..)))).....-...........---------- ( -26.50, z-score = -2.44, R) >droSim1.chr3L 4112212 93 - 22553184 CCUGUCAGCAGAAUCCACAAAACAACUCAGAUUCCGGCUGUGUUCAGCCAAGCUUCUCUUGCGGAUUCCGCCAACAGGGCCC-AAAAUCUGUAU---------- (((((..((.((((((...................(((((....)))))..((.......)))))))).))..)))))....-...........---------- ( -30.20, z-score = -3.65, R) >droSec1.super_2 4562593 93 - 7591821 CCUGUCAGCAGAAUCCACAAAACAACUCAGAUUCCGGCUGUGUUCAGCCAAGCUUCUCUUGCGGAUUCCGCCAACAGGGCCC-AAAAUCUGUAU---------- (((((..((.((((((...................(((((....)))))..((.......)))))))).))..)))))....-...........---------- ( -30.20, z-score = -3.65, R) >droYak2.chr3L 5154739 94 - 24197627 CCUGUCAGCAGUAUCCACAAAACAACUCAGAUUCCGGCUGUGCUCAGCCAAGCUUCUCUUGCGGAUUCCGCCAACAGGGCCCAAAAAUCUGUAU---------- (((((..((.(.((((...................(((((....)))))..((.......)))))).).))..)))))................---------- ( -25.80, z-score = -2.10, R) >droEre2.scaffold_4784 7290366 93 - 25762168 CCUGUCAGCAGAAUCCACAAAACAACUCAGAUUCCGGCUGUGCUCAGCCAAGCUUCUCUUGCGGAUUCCGCCAACCGGGCUC-AAAUUCUGUAU---------- .......(((((((...............((.((((((((....)))))..((.......))))).)).(((.....)))..-..)))))))..---------- ( -26.40, z-score = -2.17, R) >droAna3.scaffold_13337 2658497 104 - 23293914 CCUGUCAGCAGAAUCAACACAACAACUCUGAUUCCGGCAAUGUUCAGCCGAACUUCACUUGCGGAUUCCGCCAACAUAGCCCCAAAAACUCGGUGUGGCCGUGU .(((....))).....((((.........((.((((.(((.((((....)))).....))))))).)).((((.....(((..........))).)))).)))) ( -22.90, z-score = 0.17, R) >consensus CCUGUCAGCAGAAUCCACAAAACAACUCAGAUUCCGGCUGUGCUCAGCCAAGCUUCUCUUGCGGAUUCCGCCAACAGGGCCC_AAAAUCUGUAU__________ (((((..((.((((((...................(((((....)))))..((.......)))))))).))..))))).......................... (-22.52 = -24.02 + 1.50)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:03:05 2011