| Sequence ID | dm3.chr2L |
|---|---|
| Location | 4,316,405 – 4,316,508 |
| Length | 103 |
| Max. P | 0.663482 |

| Location | 4,316,405 – 4,316,508 |
|---|---|
| Length | 103 |
| Sequences | 11 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 85.34 |
| Shannon entropy | 0.31219 |
| G+C content | 0.37873 |
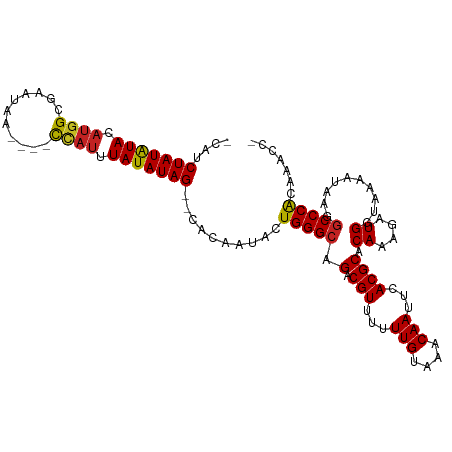
| Mean single sequence MFE | -21.13 |
| Consensus MFE | -16.29 |
| Energy contribution | -16.07 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.663482 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
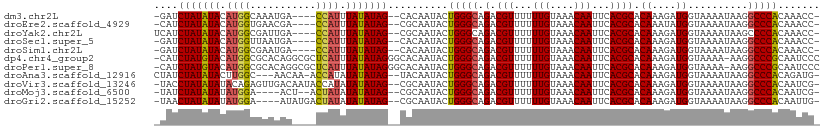
>dm3.chr2L 4316405 103 + 23011544 -GAUCUAUAUACAUGGCAAAUGA----CCAUUUAUAUAG--CACAAUACUGGGCAGACGUUUUUUGUAAACAAUUCACGCACAAAGAUGGUAAAAUAAGGCCCACAAACC- -...(((((((.(((((....).----)))).)))))))--........(((((.(.(((...(((....)))...)))).((....))..........)))))......- ( -19.90, z-score = -1.60, R) >droEre2.scaffold_4929 4393265 103 + 26641161 -CAUCUAUAUACAUGGUGAACGA----CCAUUUAUAUAG--CGCAAUACUGGGCAGACGUUUUUUGUAAACAAUUCACGCACAAAUAUGGUAAAAUAAGGCCCACAAACC- -...(((((((.(((((.....)----)))).)))))))--........(((((.(.(((...(((....)))...)))).....(((......)))..)))))......- ( -21.80, z-score = -1.73, R) >droYak2.chr2L 4347469 104 + 22324452 UCAUCUAUAUACAUGGCGAUUGA----CCAUUUAUAUAG--CGCAAUACUGGGCAGACGUUUUUUGUAAACAAUUCACGCACAAAGAUGGUAAAAUAAGCCCCACAAACC- ..............(((.....(----((((((.....(--(.((....)).)).(.(((...(((....)))...))))....))))))).......))).........- ( -18.50, z-score = -0.64, R) >droSec1.super_5 2414091 103 + 5866729 -GAUCUAUAUACAUGGUUAAUGA----CCAUUUAUAUAG--CACAAUACUGGGCAGACGUUUUUUGUAAACAAUUCACGCACAAAGAUGGUAAAAUAAGGCCCACAAACC- -...(((((((.((((((...))----)))).)))))))--........(((((.(.(((...(((....)))...)))).((....))..........)))))......- ( -21.40, z-score = -1.95, R) >droSim1.chr2L 4238670 103 + 22036055 -GAUCUAUAUACAUGGCGAAUGA----CCAUUUAUAUAG--CACAAUACUGGGCAGACGUUUUUUGUAAACAAUUCACGCACAAAGAUGGUAAAAUAAGGCCCACAAACC- -...(((((((.((((.......----)))).)))))))--........(((((.(.(((...(((....)))...)))).((....))..........)))))......- ( -19.30, z-score = -1.25, R) >dp4.chr4_group2 311746 109 + 1235136 -CAUCUAUGUACAUGGCGCACAGGCGCUCAUUUAUAUAGGGCACAAUACUGGGCAGACGUUUUUUGUAAACAAUUCACGCACAAAGAUGGUAAAA-AAGGCCCGCAAUCCC -..((((((((.(((((((....)))).))).))))))))..........((((..((..(((((((.............)))))))..))....-...))))........ ( -26.62, z-score = -1.07, R) >droPer1.super_8 388329 109 - 3966273 -CAUCUAUGUACAUGGCGCACAGGCGCUCAUUUAUAUAGGGCACAAUACUGGGCAGACGUUUUUUGUAAACAAUUCACGCACAAAGAUGGUAAAA-AAGGCCCGCAAUCCC -..((((((((.(((((((....)))).))).))))))))..........((((..((..(((((((.............)))))))..))....-...))))........ ( -26.62, z-score = -1.07, R) >droAna3.scaffold_12916 1512901 104 + 16180835 CUAUCUAUAUACUUGGC---AACAA-ACCAUAUAUAUAG--UACAAUACUGGGCAGACGUUUUUUGUAAACAAUUCACGCACAAAGAUGGUAAAAUAAGGCCCACAGAUG- ....(((((((..(((.---.....-.)))..)))))))--........(((((.(.(((...(((....)))...)))).((....))..........)))))......- ( -18.70, z-score = -1.03, R) >droVir3.scaffold_13246 386444 107 + 2672878 -UACCUAUAUAUACAGAGUUGACAAUACCAUAUAUAUAG--CGCAAUACUGGGCAGACGUUUUUUGUAAACAAUUCACGCACAAAGAUGGUAAAAUAAGGCCCACAAUCG- -...(((((((((..(.((.......))).)))))))))--........(((((.(.(((...(((....)))...)))).((....))..........)))))......- ( -15.60, z-score = 0.12, R) >droMoj3.scaffold_6500 386687 101 + 32352404 -UAUCUAUAUAUAUGGA----ACU--ACUAUAUAUAUAG--CGCAAUACUGGGCAGACGUUUUUUGUAAACAAUUCACGCACAAAGAUGGUAAAAUAAGGCCCACAAUCG- -...((((((((((((.----...--.))))))))))))--........(((((.(.(((...(((....)))...)))).((....))..........)))))......- ( -21.40, z-score = -2.36, R) >droGri2.scaffold_15252 334736 103 - 17193109 -UAACUAUAUAUAUGGA----AUAUGACUAUAUAUAUAG--CGCAAUACUGGGCAGACGUUUUUUGUAAACAAUUCACGCACAAAGAUGGUAAAAUAAGGCCCACAAUUG- -...((((((((((((.----......))))))))))))--..((((..(((((.(.(((...(((....)))...)))).((....))..........)))))..))))- ( -22.60, z-score = -2.52, R) >consensus _CAUCUAUAUACAUGGCGAAUAA____CCAUUUAUAUAG__CACAAUACUGGGCAGACGUUUUUUGUAAACAAUUCACGCACAAAGAUGGUAAAAUAAGGCCCACAAACC_ ....(((((((.((((...........)))).)))))))..........(((((.(.(((...(((....)))...)))).((....))..........)))))....... (-16.29 = -16.07 + -0.21)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:16:04 2011