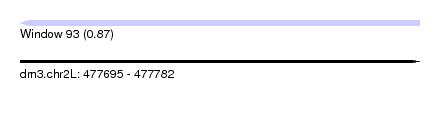
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 477,695 – 477,782 |
| Length | 87 |
| Max. P | 0.866833 |

| Location | 477,695 – 477,782 |
|---|---|
| Length | 87 |
| Sequences | 10 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 54.05 |
| Shannon entropy | 0.95076 |
| G+C content | 0.59907 |
| Mean single sequence MFE | -22.23 |
| Consensus MFE | -7.97 |
| Energy contribution | -7.93 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.86 |
| Mean z-score | -0.94 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.866833 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
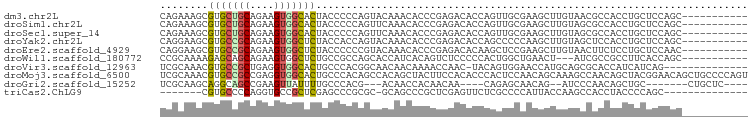
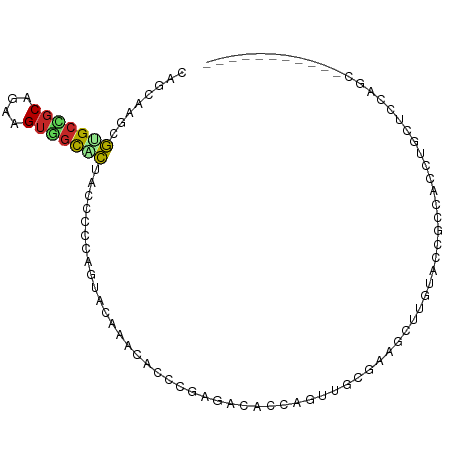
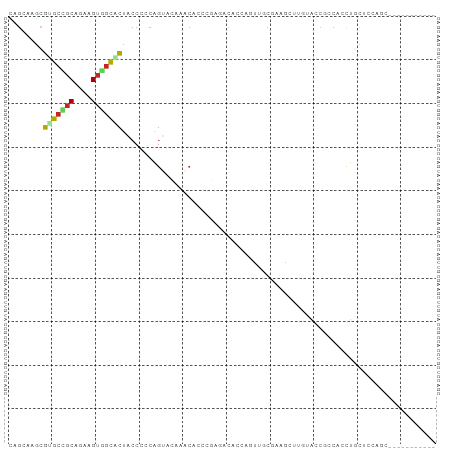
>dm3.chr2L 477695 87 - 23011544 CAGAAAGCGUGCUGCAGAAGUGGCACUACCCCCAGUACAAACACCCGAGACACCAGUUGCGAAGCUUGUAACGCCACCUGCUCCAGC----------- ......((.((..((((..((((((((......)))...................(((((((...))))))))))))))))..))))----------- ( -21.60, z-score = -0.60, R) >droSim1.chr2L 484065 87 - 22036055 CAGAAAGCGUGCUGCAGAAGUGGCACUACCCCCAGUUCAAACACCCGAGACACCAGUUGCGAAGCUUGUAGCGCCACCUGCUCCAGC----------- ......((.((..((((..(((((.((((.....((....))...((..((....))..))......)))).)))))))))..))))----------- ( -22.70, z-score = -0.38, R) >droSec1.super_14 460635 87 - 2068291 CAGAAAGCGUGCUGCAGAAGUGGCACUACCCCCAGUUCAAACACCCGAGACACCAGUUGCGAAGCUUGUAGCGCCACCUGCUCCAGC----------- ......((.((..((((..(((((.((((.....((....))...((..((....))..))......)))).)))))))))..))))----------- ( -22.70, z-score = -0.38, R) >droYak2.chr2L 462107 87 - 22324452 CAGGAAGCGUGCCGCAGAAGUGGCUCUACCACCAGUACAAACACCCGAGACACCAGCCCCCAAGCUUGUAGCUCCACCUGCUCCAGC----------- ..(((.(((.(((((....))))).)....................(((..((.(((......))).))..))).....)))))...----------- ( -21.80, z-score = -0.58, R) >droEre2.scaffold_4929 529872 87 - 26641161 CAGGAAGCGUGCCGCAGAAGUGGCUCUACCCCCCGUACAAACACCCGAGACACAAGCUCCGAAGCUUGUAACUUCUCCUGCUCCAAC----------- ..(((.(((.(((((....))))).)....................((((.(((((((....)))))))....))))..)))))...----------- ( -25.40, z-score = -2.51, R) >droWil1.scaffold_180772 5968820 84 + 8906247 CCGCAAAAGAGCAGCAGAAGUGGCUCUGCCGCCAGCACCAUCACAGUCUCCCCCACUGGCUGAACU---AUCGCCGCCUUCACCAGC----------- ..((......))....((((((((..(((.....)))......(((((.........)))))....---...)))).))))......----------- ( -18.90, z-score = 0.50, R) >droVir3.scaffold_12963 18897239 83 - 20206255 UCGCAAACGUGCCGCUGAGGUGGCACUGCCCACGGCAACAACAAAACCAAC-UACAGUGGAACCAUGCAGCGCACCAUCAUCAG-------------- ........((((.((((.(((..(((((.....((...........))...-..)))))..)))...)))))))).........-------------- ( -22.00, z-score = -0.41, R) >droMoj3.scaffold_6500 8071924 98 + 32352404 UCGCAAACGUGCCGCCGAGGUGGCACUGCCCACAGCCACAGCUACUUCCACACCCACUCCAACAGCAAAGCCAACAGCUACGGAACAGCUGCCCCAGU ..(((...(((((((....)))))))))).........(((((..((((.......((.....))...(((.....)))..)))).)))))....... ( -24.90, z-score = -0.50, R) >droGri2.scaffold_15252 6804914 78 - 17193109 UCGCAAGCAGGCAGCCGAAGUUAUUUUGCCCACG---ACAACCACAACAA----CAGAGCAACAG--AUCCCAACAGCUGC-------CUGCUC---- .....((((((((((.(..(..(((((((.(...---.............----..).))))..)--))..)..).)))))-------))))).---- ( -21.47, z-score = -3.32, R) >triCas2.ChLG9 13723635 76 - 15222296 -------CGUGCCCCAGGUGCCGCUCGAGCCCGCGC-GCAGCCCGCUCGAGUUCUCGCCCCAUUACCAAGCCACCUACCCCAGC-------------- -------.........((((..((((((((..((..-...))..))))))))...)))).........................-------------- ( -20.80, z-score = -1.24, R) >consensus CAGCAAGCGUGCCGCAGAAGUGGCACUACCCCCAGUACAAACACCCGAGACACCAGUUGCGAAGCUUGUACCGCCACCUGCUCCAGC___________ ........(((((((....)))))))........................................................................ ( -7.97 = -7.93 + -0.04)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:05:56 2011