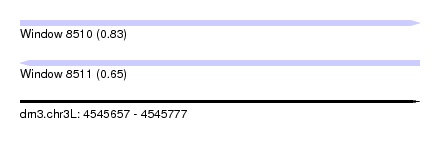
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 4,545,657 – 4,545,777 |
| Length | 120 |
| Max. P | 0.830688 |

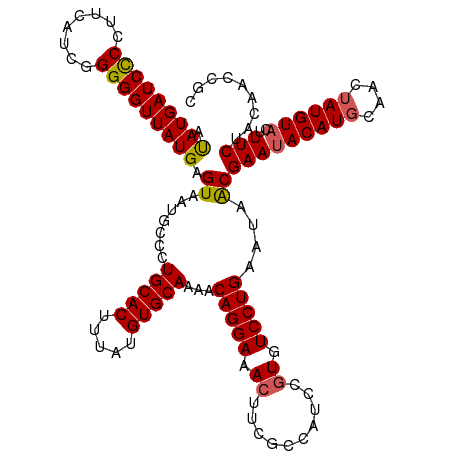
| Location | 4,545,657 – 4,545,777 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.66 |
| Shannon entropy | 0.11143 |
| G+C content | 0.45472 |
| Mean single sequence MFE | -30.02 |
| Consensus MFE | -27.30 |
| Energy contribution | -27.74 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.830688 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 4545657 120 + 24543557 AUAUGAUCCCCUCCAUCGGGGGUUAUGAGUAAUGCCCUGCACUUUAUGUGCAAAACAGGAAACUUCGCCAUCCGUGUCCUGAAUAACGAAUACAUGCAACUAUGUACUUUCUACAACCGC .((((((((((......)))))))))).(((......(((((.....)))))...(((((.((..........)).)))))......(((((((((....))))))..))))))...... ( -31.40, z-score = -2.34, R) >droSim1.chr3L 4077259 120 + 22553184 AUAUGAUCCCCUUCAUCGGGGGUUAUGAGUAAUGCCCUGCACUUUAUGUGCAAAACAGGAAACUUCGCCAUCCGUGUCCUGAAUGACGAAUACAUGCAACGAUGUACUUUCUACAACCGC .((((((((((......)))))))))).(((......(((((.....)))))...(((((.((..........)).)))))......((((((((......)))))..))))))...... ( -31.10, z-score = -1.66, R) >droSec1.super_2 4528276 120 + 7591821 AUAUGAUCUCCUGCAUCGGGGGUUAUGAGUAAUGCCCUGCACUUUAUGUGCAAAACAGGAAACUUCGCCAUCCGUGUCCUGAAUAACGAAUACAUGCAACUAUGUACUUUCUACCACCGC .((((((((((......))))))))))..........(((((.....)))))...(((((.((..........)).)))))......(((((((((....))))))..)))......... ( -27.60, z-score = -0.81, R) >droYak2.chr3L 5118456 118 + 24197627 ACAUGAUCCC--UCCUUGGGGGUUAUGAGUAAUGCCCUGCACUUUAUGUGCAAAACAGGAAACUUCGACAUCCCUGUCCUGAAUAACGAAUACAUGCAACUAUGUUCUUUCUACGACCGC .(((((((((--(....)))))))))).(((......(((((.....)))))...(((((......(....)....)))))......(((.(((((....)))))...))))))...... ( -28.50, z-score = -1.75, R) >droEre2.scaffold_4784 7254193 118 + 25762168 ACAUGAUCCC--UCCUUGGGGGUUAUGAGUAAUGCCCUGCACUUUAUGUGCAAAACAGGAAACUUCGCCAUCCGUGUCCUGAAUAGCGAAUACAUGCAACUAUGUACUUUCCACAACCGC .(((((((((--(....))))))))))..........(((((.....))))).....(((((.(((((.((.((.....)).)).)))))((((((....)))))).)))))........ ( -31.50, z-score = -1.85, R) >consensus AUAUGAUCCCCUUCAUCGGGGGUUAUGAGUAAUGCCCUGCACUUUAUGUGCAAAACAGGAAACUUCGCCAUCCGUGUCCUGAAUAACGAAUACAUGCAACUAUGUACUUUCUACAACCGC .((((((((((......)))))))))).((.......(((((.....)))))...(((((.((..........)).)))))....))(((((((((....))))))..)))......... (-27.30 = -27.74 + 0.44)

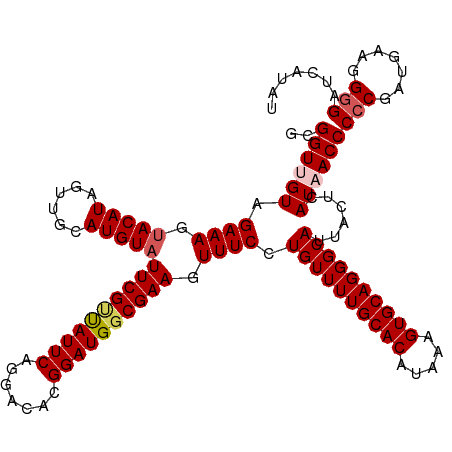
| Location | 4,545,657 – 4,545,777 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.66 |
| Shannon entropy | 0.11143 |
| G+C content | 0.45472 |
| Mean single sequence MFE | -35.00 |
| Consensus MFE | -31.12 |
| Energy contribution | -31.84 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.651012 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 4545657 120 - 24543557 GCGGUUGUAGAAAGUACAUAGUUGCAUGUAUUCGUUAUUCAGGACACGGAUGGCGAAGUUUCCUGUUUUGCACAUAAAGUGCAGGGCAUUACUCAUAACCCCCGAUGGAGGGGAUCAUAU ..((((((.((((.(((((......)))))((((((((((.......)))))))))).)))).((((((((((.....))))))))))......))))))(((......)))........ ( -35.70, z-score = -1.69, R) >droSim1.chr3L 4077259 120 - 22553184 GCGGUUGUAGAAAGUACAUCGUUGCAUGUAUUCGUCAUUCAGGACACGGAUGGCGAAGUUUCCUGUUUUGCACAUAAAGUGCAGGGCAUUACUCAUAACCCCCGAUGAAGGGGAUCAUAU ..((((((.((((.(((((......)))))((((((((((.......)))))))))).)))).((((((((((.....))))))))))......))))))(((......)))........ ( -37.60, z-score = -2.15, R) >droSec1.super_2 4528276 120 - 7591821 GCGGUGGUAGAAAGUACAUAGUUGCAUGUAUUCGUUAUUCAGGACACGGAUGGCGAAGUUUCCUGUUUUGCACAUAAAGUGCAGGGCAUUACUCAUAACCCCCGAUGCAGGAGAUCAUAU ((..(.(((.....)))...)..)).....((((((((((.......))))))))))(((((((((..(((((.....)))))(((..(((....)))..)))...)))))))))..... ( -34.00, z-score = -1.33, R) >droYak2.chr3L 5118456 118 - 24197627 GCGGUCGUAGAAAGAACAUAGUUGCAUGUAUUCGUUAUUCAGGACAGGGAUGUCGAAGUUUCCUGUUUUGCACAUAAAGUGCAGGGCAUUACUCAUAACCCCCAAGGA--GGGAUCAUGU ..(((((((((((..((((......)))).((((.(((((.......))))).)))).)))).((((((((((.....)))))))))).)))......((((...)).--)))))).... ( -27.10, z-score = 0.36, R) >droEre2.scaffold_4784 7254193 118 - 25762168 GCGGUUGUGGAAAGUACAUAGUUGCAUGUAUUCGCUAUUCAGGACACGGAUGGCGAAGUUUCCUGUUUUGCACAUAAAGUGCAGGGCAUUACUCAUAACCCCCAAGGA--GGGAUCAUGU ..(((((((((((.(((((......)))))((((((((((.......)))))))))).)))))((((((((((.....))))))))))......))))))(((.....--)))....... ( -40.60, z-score = -2.75, R) >consensus GCGGUUGUAGAAAGUACAUAGUUGCAUGUAUUCGUUAUUCAGGACACGGAUGGCGAAGUUUCCUGUUUUGCACAUAAAGUGCAGGGCAUUACUCAUAACCCCCGAUGAAGGGGAUCAUAU ..((((((.((((.(((((......)))))((((((((((.......)))))))))).)))).((((((((((.....))))))))))......))))))(((.......)))....... (-31.12 = -31.84 + 0.72)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:02:55 2011