| Sequence ID | dm3.chr3L |
|---|---|
| Location | 4,524,296 – 4,524,387 |
| Length | 91 |
| Max. P | 0.647502 |

| Location | 4,524,296 – 4,524,387 |
|---|---|
| Length | 91 |
| Sequences | 9 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 80.43 |
| Shannon entropy | 0.38606 |
| G+C content | 0.38046 |
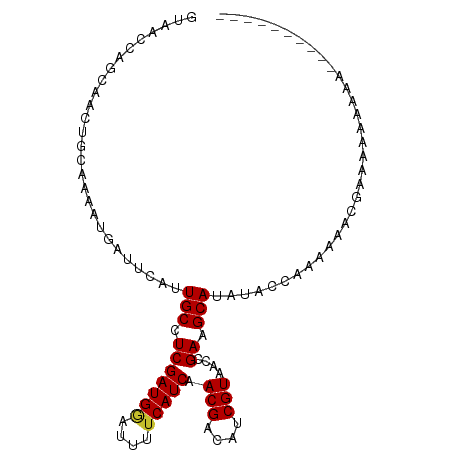
| Mean single sequence MFE | -14.31 |
| Consensus MFE | -9.94 |
| Energy contribution | -9.87 |
| Covariance contribution | -0.07 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.647502 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
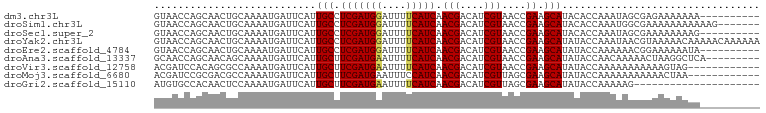
>dm3.chr3L 4524296 91 + 24543557 GUAACCAGCAACUGCAAAAUGAUUCAUUGCCUCGAUGGAUUUUCAUCAACGACAUCGUAACCGAAGCAUACACCAAAUAGCGAGAAAAAAA---------- (((....((....))............(((.(((((((....))))).(((....)))....)).))))))....................---------- ( -12.60, z-score = -0.20, R) >droSim1.chr3L 4056217 94 + 22553184 GUAACCAGCAACUGCAAAAUGAUUCAUUGCCUCGAUGGAUUUUCAUCAACGACAUCGUAACCGAAGCAUACACCAAAUGGCGAAAAAAAAAAAG------- ....(((((....))............(((.(((((((....))))).(((....)))....)).))).........)))..............------- ( -13.70, z-score = -0.28, R) >droSec1.super_2 4507332 91 + 7591821 GUAACCAGCAACUGCAAAAUGAUUCAUUGCCUCGAUGGAUUUUCAUCAACGACAUCGUAACCGAAGCAUACACCAAAUAGCGAAAAAAAAG---------- (((....((....))............(((.(((((((....))))).(((....)))....)).))))))....................---------- ( -12.60, z-score = -0.55, R) >droYak2.chr3L 5096063 101 + 24197627 GUAACCAGCAACUGCAAAAUGAUUCAUUGCCUCGAUGGAUUUUCAUCAACGACAUCGUAACCGAAGCAUAUACCAAAUAACGUAAAAACAAAAACAAAAAA (((....((....))............(((.(((((((....))))).(((....)))....)).)))..)))............................ ( -11.70, z-score = -0.54, R) >droEre2.scaffold_4784 7230975 91 + 25762168 GUAACCAGCAACUGCAAAAUGAUUCAUUGCCUCGAUGGAUUUUCAUCAACGACAUCGUAACCGAAGCAUAUACCAAAAAACGGAAAAAAUA---------- ....((.((....))............(((.(((((((....))))).(((....)))....)).))).............))........---------- ( -13.70, z-score = -0.88, R) >droAna3.scaffold_13337 2605838 92 + 23293914 GCAACCAGCAACAGCAAAAUGAUUCAUUGCUUCGAUGAAUUUUCAUCAACGACAUCGUAACCGAAGCAUAUACCAACAAAAACUAAGGCUCA--------- ((.....((....))............(((((((((((....))))).(((....)))....))))))...................))...--------- ( -17.80, z-score = -2.89, R) >droVir3.scaffold_12758 98966 89 + 972128 ACGAUCCACAGCGCCAAAAUGAUUCAUUGCUUCGAUGAAUUUUCAUCAACGACAUCGUAACCGAAGCAUAUACCAAAAAAAAAAAGUAG------------ ...........................(((((((((((....))))).(((....)))....)))))).....................------------ ( -14.90, z-score = -2.20, R) >droMoj3.scaffold_6680 1224832 89 + 24764193 ACGAUCCGCGACGCCAAAAUGAUUCAUUGCUUCGAUGAAUUUCCAUCAACGACAUCGUUAGCGAAGCAUAUACCAAAAAAAAAAACUAA------------ .......((..(((......(((((((((...)))))))))......((((....)))).)))..))......................------------ ( -16.00, z-score = -1.78, R) >droGri2.scaffold_15110 23302829 80 - 24565398 AUGUGCCACAACUCCAAAAUGAUUCAUUGCUUCGAUGAAUUUUCAUCAACGACAUCGUUAGCGAAGCAUAUACCAAAAAG--------------------- ...........................(((((((((((....)))))((((....))))...))))))............--------------------- ( -15.80, z-score = -2.28, R) >consensus GUAACCAGCAACUGCAAAAUGAUUCAUUGCCUCGAUGGAUUUUCAUCAACGACAUCGUAACCGAAGCAUAUACCAAAAAACGAAAAAAAAA__________ ...........................(((.(((((((....))))).(((....)))....)).)))................................. ( -9.94 = -9.87 + -0.07)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:02:53 2011