| Sequence ID | dm3.chr3L |
|---|---|
| Location | 4,517,832 – 4,517,931 |
| Length | 99 |
| Max. P | 0.757584 |

| Location | 4,517,832 – 4,517,931 |
|---|---|
| Length | 99 |
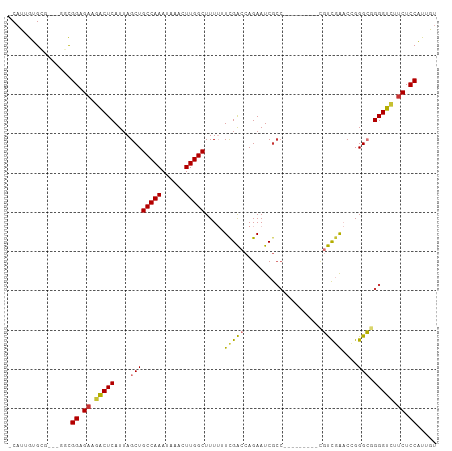
| Sequences | 8 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 78.78 |
| Shannon entropy | 0.38964 |
| G+C content | 0.53662 |
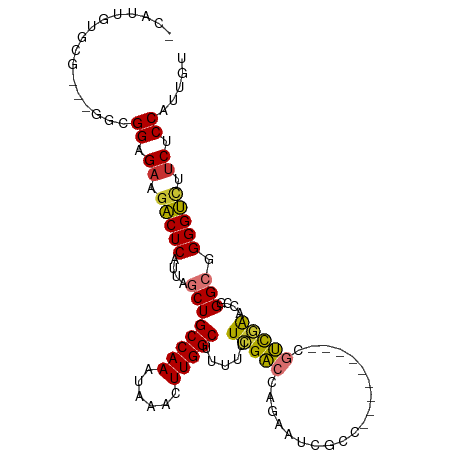
| Mean single sequence MFE | -34.79 |
| Consensus MFE | -21.47 |
| Energy contribution | -20.56 |
| Covariance contribution | -0.91 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.757584 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 4517832 99 - 24543557 -CAUUGUGUGGCUGGCGGAGAAGACUCAUUAGCUGCCAAAUAAACUUGGCUUUUUUCGACCAGAAUCGCC---------CGUCGAACGGGGCGGGGUCUUCUCCAUUGU -...............(((((((((((.......(((((......)))))................((((---------(........))))))))))))))))..... ( -36.30, z-score = -2.08, R) >droSim1.chr3L 4048462 98 - 22553184 -CAUUGUGUGGCUGGCGGAGAAGACUCAUUAGCUGCCAAAUAAACUUGGCUUUU-UCGACCAGAAUCGCC---------CGUCGAACCGGGCGGGGUCUUCUCCAUUGU -...............(((((((((((.......(((((......)))))....-...........((((---------((......)))))))))))))))))..... ( -38.30, z-score = -2.79, R) >droSec1.super_2 4500908 99 - 7591821 -CAUUGUGUGGCUGGCGGAGUAGACUCAUUAGCUGCCAAAUAAACUUGGCUUUUAUCGACCAGAAUCGCC---------CGUCGAACCGGGCGGGGUCUUCUCCAUUGU -...............((((.((((((..(((..(((((......)))))..)))...........((((---------((......)))))))))))).))))..... ( -34.00, z-score = -1.61, R) >droYak2.chr3L 5089464 99 - 24197627 -CAUUGUGCGGCUGGCGGCGAAGACUCAUUAGCUGCCAAAUAAACUUGGCUUUUUUCGACCAGAAUCGCC---------CGUCGAACCGGGCGGGGUCUUCUCCAUUGU -......((.....))((.((((((((.......(((((......)))))................((((---------((......)))))))))))))).))..... ( -34.90, z-score = -1.40, R) >droEre2.scaffold_4784 7222889 96 - 25762168 -CAUUGUGCG---GCUGGCGAAGACUCAUUAGCUGCCAAAUAAACUUGGCUUUUUUCGACCAGAAUCGCC---------CGUCGAACCGGGCGGGGUCGUCUCCAUUGU -..(((.(((---(((((.((....)).))))))))))).......(((.......(((((....(((((---------((......))))))))))))...))).... ( -33.20, z-score = -1.43, R) >droAna3.scaffold_13337 2599165 95 - 23293914 -CUUUGUGC----GGCGGCGAAGACUCAUUAGCUGCCAAAUAAACUUGGCUUUUUUCGGCCAGAAUCAAG---------AGCUGAACCGGGCUGGGUAUUCUCCAUUGU -..((((..----((((((............))))))..))))(((..((((..(((((((........)---------.))))))..))))..)))............ ( -29.00, z-score = -0.74, R) >dp4.chrXR_group8 2616856 100 - 9212921 GCAUUGUG-------CGGUGAUGCCUCAUUAGCUGCCAAAUAAACUUGGCUUU--UCGGUCAGAAUCGCCGCCAGUCCGUCCCAGUCUGGGGUGGGGUCUCUCCGCCAU ..((((.(-------(((((((..((.((..(..(((((......)))))...--)..)).)).))))))))))))((..(((.....)))..))(((......))).. ( -35.50, z-score = -1.35, R) >droPer1.super_24 1122778 100 + 1556852 GCAUUGUG-------CGGUGAUGCCUCAUUAGCUGCCAAAUAAACUUGGCUUU--UUGGUCAGAAUCGCCGCCAGUCCGUCCCAGUCUGGGGUGGGGUCUCUCCGCCAU ..((((.(-------(((((((..((.(((((..(((((......)))))...--))))).)).))))))))))))((..(((.....)))..))(((......))).. ( -37.10, z-score = -1.82, R) >consensus _CAUUGUGCG___GGCGGAGAAGACUCAUUAGCUGCCAAAUAAACUUGGCUUUUUUCGACCAGAAUCGCC_________CGUCGAACCGGGCGGGGUCUUCUCCAUUGU ................((.((.(((((.......(((((......))))).............................((((......))))))))).)).))..... (-21.47 = -20.56 + -0.91)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:02:52 2011