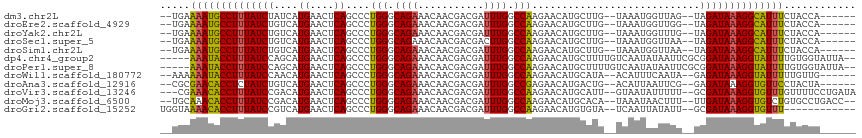
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 4,316,279 – 4,316,383 |
| Length | 104 |
| Max. P | 0.999369 |

| Location | 4,316,279 – 4,316,383 |
|---|---|
| Length | 104 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 78.71 |
| Shannon entropy | 0.45667 |
| G+C content | 0.41254 |
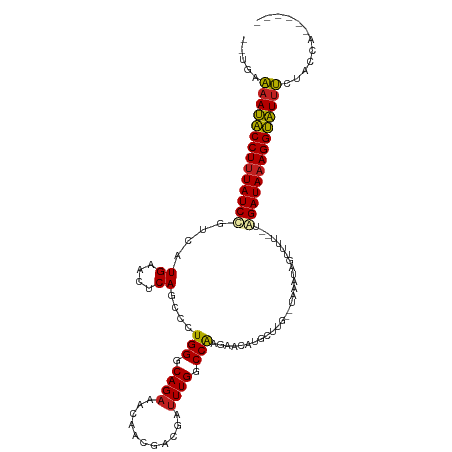
| Mean single sequence MFE | -34.36 |
| Consensus MFE | -30.27 |
| Energy contribution | -28.99 |
| Covariance contribution | -1.28 |
| Combinations/Pair | 1.29 |
| Mean z-score | -3.83 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.83 |
| SVM RNA-class probability | 0.999369 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
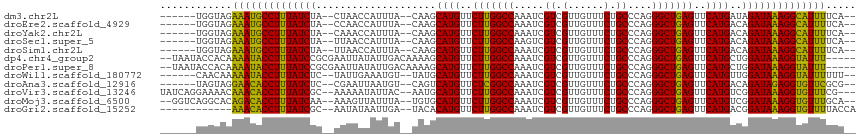
>dm3.chr2L 4316279 104 + 23011544 ------UGGUAGAAAUGCCUUUAUCUA--CUAACCAUUUA--CAAGCAUGUUCUUGGCCAAAUCGUCGUUGUUUCUGCCCAGGGCUGAGUUCAUGAUAGAUAAAGGCAUUUUCA-- ------.....((((((((((((((((--...........--....((((..((..(((.....((.(......).))....)))..))..)))).))))))))))))))))..-- ( -33.19, z-score = -3.51, R) >droEre2.scaffold_4929 4393143 104 + 26641161 ------UGGUAGAAAUGCCUUUAUCUA--CCAACCAUUUA--CAAGCAUGUUCUUGGCCAAAUCGUCGUUGUUUCUGCCCAGGGCUGAGUUCAUGACAGAUAAAGGCAUUUUCA-- ------.....(((((((((((((((.--....(((...(--((....)))...))).......(((((...(((.(((...))).)))...))))))))))))))))))))..-- ( -33.20, z-score = -3.31, R) >droYak2.chr2L 4347345 104 + 22324452 ------UGGUAGAAAUGCCUUUAUCUA--CAAACCAUUUA--CAAGCAUGUUCUUGGCCAAAUCGUCGUUGUUUCUGCCCAGGGCUGAGUUCAUGACAGAUAAAGGCAUUUUCA-- ------.....(((((((((((((((.--....(((...(--((....)))...))).......(((((...(((.(((...))).)))...))))))))))))))))))))..-- ( -33.20, z-score = -3.39, R) >droSec1.super_5 2413967 104 + 5866729 ------UGGUAGAAAUGCCUUUAUCUA--UUAACCAUUUA--CAAGCAUGUUCUUGGCCAAGUCGUCGUUGUUUCUGCCCAGGGCUGAGUUCAUGACAGAUAAAGGCAUUUUCA-- ------.....(((((((((((((((.--....(((...(--((....)))...))).......(((((...(((.(((...))).)))...))))))))))))))))))))..-- ( -33.20, z-score = -3.32, R) >droSim1.chr2L 4238546 104 + 22036055 ------UGGUAGAAAUGCCUUUAUCUA--UUAACCAUUUA--CAAGCAUGUUCUUGGCCAAAUCGUCGUUGUUUCUGCCCAGGGCUGAGUUCAUGACAGAUAAAGGCAUUUUCA-- ------.....(((((((((((((((.--....(((...(--((....)))...))).......(((((...(((.(((...))).)))...))))))))))))))))))))..-- ( -33.20, z-score = -3.51, R) >dp4.chr4_group2 311613 109 + 1235136 --UAAUACCACAAAAUACCUUUAUCCGCGAAUUAUAUUGACAAAAGCAUGUUCUUGGCCAAAUCGUCGUUGUUUCUGCCCAGGGCUGAGUUCAUGCUGGAUAAAGGUAUUU----- --..........((((((((((((((((((......))).)...((((((..((..(((.....((.(......).))....)))..))..))))))))))))))))))))----- ( -37.60, z-score = -5.01, R) >droPer1.super_8 388196 109 - 3966273 --UAAUACCACAAAAUACCUUUAUCCGCGAAUUAUAUUGACAAAAGCAUGUUCUUGGCCAAAUCGUCGUUGUUUCUGCCCAGGGCUGAGUUCAUGCUGGAUAAAGGUAUUU----- --..........((((((((((((((((((......))).)...((((((..((..(((.....((.(......).))....)))..))..))))))))))))))))))))----- ( -37.60, z-score = -5.01, R) >droWil1.scaffold_180772 2040151 104 + 8906247 ------CAACAAAAAUACCUUUAUCUC--UAUUGAAAUGU--UAUGCAUGUUCUUGGCCAAAUCGUCGUUGUUUCUGCCCAGGGCUGAGUUCAUGUUGGAUAAAGGUAUUUUUU-- ------....(((((((((((((((((--....)).....--...(((((..((..(((.....((.(......).))....)))..))..)))))..))))))))))))))).-- ( -32.00, z-score = -4.31, R) >droAna3.scaffold_12916 1512785 104 + 16180835 ------UAGUAGGAACACCUUUAUCUC--CGAAUUAAUGU--CAGUCAUGUUCUCGGCCAAAUCGUCGUUGUUUCUGCCCAGGGCUGAGUUCAUGACAGAUAGAGGUGUUCGCG-- ------......((((((((((((((.--.((.......)--).((((((..(((((((.....((.(......).))....)))))))..))))))))))))))))))))...-- ( -42.50, z-score = -5.76, R) >droVir3.scaffold_13246 386312 109 + 2672878 UAUCAGGAAAACAAACACCUUUAUCGC--AAAAAUAUUAC--AAUGCAUGUUCUUGGCCAAAUCGUCGUUGUUUCUGCCCAGGGCUGAGUUCAUGUCGGAUAAAGGUGUUUCG--- ............(((((((((((((((--(..........--..)))(((..((..(((.....((.(......).))....)))..))..)))....)))))))))))))..--- ( -30.60, z-score = -2.78, R) >droMoj3.scaffold_6500 386557 108 + 32352404 --GGUCAGGCACAGACACCUUUAUCAA--AAAGUUAUUUA--UGUGCAUGUUCUUGGCCAAAUCGUCGUUGUUUCUGCCCAGGGCUGAGUUCAUGUCGGAUAAAGGUGUUUGCA-- --.........((((((((((((((..--(((....))).--...(((((..((..(((.....((.(......).))....)))..))..)))))..))))))))))))))..-- ( -34.40, z-score = -2.56, R) >droGri2.scaffold_15252 334607 100 - 17193109 ------------AAACACCUUUAUCGC--AAUAUAAUUGA--UACACAUGUUCUUGGCCAAAUCGUCGUUGUUUCUGCCCAGGGCUGAGUUCAUGACGGAUAAAGGUGUUUUACCA ------------(((((((((((((.(--(((...)))).--....((((..((..(((.....((.(......).))....)))..))..))))...)))))))))))))..... ( -31.60, z-score = -3.46, R) >consensus ______UGGUAGAAAUACCUUUAUCUA__AAAACCAUUUA__CAAGCAUGUUCUUGGCCAAAUCGUCGUUGUUUCUGCCCAGGGCUGAGUUCAUGACAGAUAAAGGUAUUUUCA__ ............((((((((((((((....................((((..(((((((.....((.(......).))....)))))))..))))..))))))))))))))..... (-30.27 = -28.99 + -1.28)
| Location | 4,316,279 – 4,316,383 |
|---|---|
| Length | 104 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 78.71 |
| Shannon entropy | 0.45667 |
| G+C content | 0.41254 |
| Mean single sequence MFE | -28.42 |
| Consensus MFE | -22.48 |
| Energy contribution | -21.73 |
| Covariance contribution | -0.74 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.05 |
| SVM RNA-class probability | 0.997144 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
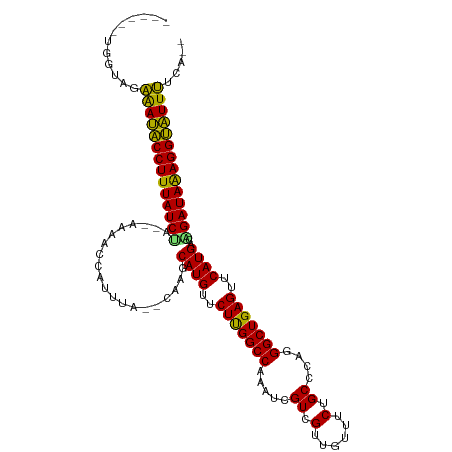
>dm3.chr2L 4316279 104 - 23011544 --UGAAAAUGCCUUUAUCUAUCAUGAACUCAGCCCUGGGCAGAAACAACGACGAUUUGGCCAAGAACAUGCUUG--UAAAUGGUUAG--UAGAUAAAGGCAUUUCUACCA------ --...((((((((((((((((..........(((...))).........(((.(((((..((((......))))--))))).))).)--)))))))))))))))......------ ( -29.90, z-score = -2.93, R) >droEre2.scaffold_4929 4393143 104 - 26641161 --UGAAAAUGCCUUUAUCUGUCAUGAACUCAGCCCUGGGCAGAAACAACGACGAUUUGGCCAAGAACAUGCUUG--UAAAUGGUUGG--UAGAUAAAGGCAUUUCUACCA------ --...(((((((((((((((((.((....))(((...))).........)))...(..((((...(((....))--)...))))..)--.))))))))))))))......------ ( -31.50, z-score = -2.68, R) >droYak2.chr2L 4347345 104 - 22324452 --UGAAAAUGCCUUUAUCUGUCAUGAACUCAGCCCUGGGCAGAAACAACGACGAUUUGGCCAAGAACAUGCUUG--UAAAUGGUUUG--UAGAUAAAGGCAUUUCUACCA------ --...(((((((((((((((.((........(((...))).........(((.(((((..((((......))))--))))).)))))--)))))))))))))))......------ ( -28.80, z-score = -2.12, R) >droSec1.super_5 2413967 104 - 5866729 --UGAAAAUGCCUUUAUCUGUCAUGAACUCAGCCCUGGGCAGAAACAACGACGACUUGGCCAAGAACAUGCUUG--UAAAUGGUUAA--UAGAUAAAGGCAUUUCUACCA------ --...(((((((((((((((((.((....))(((...))).........)))...(((((((...(((....))--)...)))))))--.))))))))))))))......------ ( -30.90, z-score = -3.33, R) >droSim1.chr2L 4238546 104 - 22036055 --UGAAAAUGCCUUUAUCUGUCAUGAACUCAGCCCUGGGCAGAAACAACGACGAUUUGGCCAAGAACAUGCUUG--UAAAUGGUUAA--UAGAUAAAGGCAUUUCUACCA------ --...(((((((((((((((((.((....))(((...))).........)))...(((((((...(((....))--)...)))))))--.))))))))))))))......------ ( -31.00, z-score = -3.30, R) >dp4.chr4_group2 311613 109 - 1235136 -----AAAUACCUUUAUCCAGCAUGAACUCAGCCCUGGGCAGAAACAACGACGAUUUGGCCAAGAACAUGCUUUUGUCAAUAUAAUUCGCGGAUAAAGGUAUUUUGUGGUAUUA-- -----((((((((((((((.((.........(((...))).........(((((...(((.........))).)))))..........))))))))))))))))..........-- ( -30.10, z-score = -2.47, R) >droPer1.super_8 388196 109 + 3966273 -----AAAUACCUUUAUCCAGCAUGAACUCAGCCCUGGGCAGAAACAACGACGAUUUGGCCAAGAACAUGCUUUUGUCAAUAUAAUUCGCGGAUAAAGGUAUUUUGUGGUAUUA-- -----((((((((((((((.((.........(((...))).........(((((...(((.........))).)))))..........))))))))))))))))..........-- ( -30.10, z-score = -2.47, R) >droWil1.scaffold_180772 2040151 104 - 8906247 --AAAAAAUACCUUUAUCCAACAUGAACUCAGCCCUGGGCAGAAACAACGACGAUUUGGCCAAGAACAUGCAUA--ACAUUUCAAUA--GAGAUAAAGGUAUUUUUGUUG------ --.((((((((((((((((................(((.((((...........)))).))).(((.(((....--.))))))....--).)))))))))))))))....------ ( -24.10, z-score = -3.02, R) >droAna3.scaffold_12916 1512785 104 - 16180835 --CGCGAACACCUCUAUCUGUCAUGAACUCAGCCCUGGGCAGAAACAACGACGAUUUGGCCGAGAACAUGACUG--ACAUUAAUUCG--GAGAUAAAGGUGUUCCUACUA------ --...((((((((.(((((.(((((..(((.(((.......................))).)))..)))))(((--(.......)))--)))))).))))))))......------ ( -29.90, z-score = -2.66, R) >droVir3.scaffold_13246 386312 109 - 2672878 ---CGAAACACCUUUAUCCGACAUGAACUCAGCCCUGGGCAGAAACAACGACGAUUUGGCCAAGAACAUGCAUU--GUAAUAUUUUU--GCGAUAAAGGUGUUUGUUUUCCUGAUA ---..(((((((((((((.(.((((....((....))(((.(((.(......).))).))).....)))))...--((((.....))--)))))))))))))))............ ( -26.50, z-score = -1.73, R) >droMoj3.scaffold_6500 386557 108 - 32352404 --UGCAAACACCUUUAUCCGACAUGAACUCAGCCCUGGGCAGAAACAACGACGAUUUGGCCAAGAACAUGCACA--UAAAUAACUUU--UUGAUAAAGGUGUCUGUGCCUGACC-- --.(((.(((((((((((.(.((((....((....))(((.(((.(......).))).))).....)))))...--.(((......)--))))))))))))).)))........-- ( -23.40, z-score = -1.05, R) >droGri2.scaffold_15252 334607 100 + 17193109 UGGUAAAACACCUUUAUCCGUCAUGAACUCAGCCCUGGGCAGAAACAACGACGAUUUGGCCAAGAACAUGUGUA--UCAAUUAUAUU--GCGAUAAAGGUGUUU------------ .....(((((((((((((((((.((....))(((...))).........))))................(((((--.....))))).--..)))))))))))))------------ ( -24.90, z-score = -1.41, R) >consensus __UGAAAAUACCUUUAUCCGUCAUGAACUCAGCCCUGGGCAGAAACAACGACGAUUUGGCCAAGAACAUGCUUG__UAAAUAGUUUU__UAGAUAAAGGUAUUUCUACCA______ .....((((((((((((((....((....))....(((.((((...........)))).)))............................))))))))))))))............ (-22.48 = -21.73 + -0.74)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:16:03 2011