
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 4,442,258 – 4,442,363 |
| Length | 105 |
| Max. P | 0.996551 |

| Location | 4,442,258 – 4,442,363 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 78.55 |
| Shannon entropy | 0.41131 |
| G+C content | 0.43343 |
| Mean single sequence MFE | -22.03 |
| Consensus MFE | -11.71 |
| Energy contribution | -12.10 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.870346 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
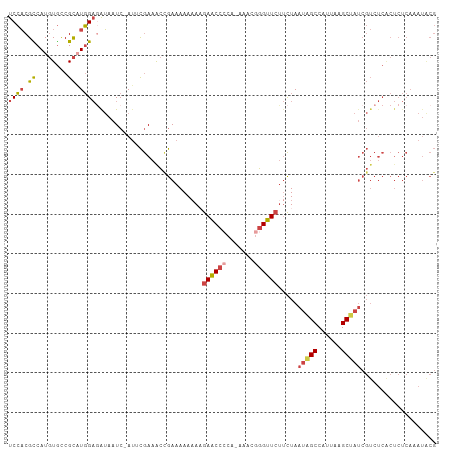
>dm3.chr3L 4442258 105 + 24543557 UCCACGCCAUGUGCCGCAUGGAAAUAAUC-AUUCGAAGCCGAAAAAAA-GAACCGCA-AAACGGGUUCUUCUAAUAGCCAUUAAGCUAUCGUCUCACUCUCAAAUACG ......(((((.....)))))........-....((.(.(((....((-(((((...-.....)))))))....((((......))))))).)))............. ( -20.10, z-score = -1.32, R) >droSim1.chr3L 3977802 106 + 22553184 UCCACGCCAUGUGCCGCAUGGAGAUAAUC-AUUCGAAACCGAAAAAAAAGAACCCCA-AAACGGGUUCUUCUAAUAGCCAUUAAGCUAUCGUCUCACUCUCAAAUGCG ....(((((((.....))))(((((....-.((((....))))....((((((((..-....))))))))...(((((......))))).)))))..........))) ( -27.60, z-score = -3.98, R) >droSec1.super_2 4431041 106 + 7591821 UCCACGCCAUGUGCCGCAUGGAGAUAAUC-AUUCGAAACCGAAAAAAAAGAACCCCA-AAACGGGUUCUUCUAAUAGCCAUUAAGCUAUCGUCUCACUCUCAAAUGCG ....(((((((.....))))(((((....-.((((....))))....((((((((..-....))))))))...(((((......))))).)))))..........))) ( -27.60, z-score = -3.98, R) >droYak2.chr3L 5019729 106 + 24197627 UCCACGCCAUGUGCCGCCUGGAGAUAAUC-AUACGAAGCUGAAAAAUAAGAACCCCA-AAACGGGUUCUUCUAAUGGCCAUUAAGCCAUCGUCUCACUCUCAAAUACG ((((.((........)).)))).......-....((..(........((((((((..-....))))))))...(((((......))))).)..))............. ( -24.00, z-score = -2.11, R) >droEre2.scaffold_4784 7150872 105 + 25762168 UCCACGUCAUGUGCCGCAUGGAGGUAAUC-AUGCGAAGCCGAAAAAAA-GAACACCA-AAACGGGUUCUUCUAAUGGCCAUUAAGCCAUCGUCUCGCUCUCAAAUACG ....(..((((.....))))..)(((...-..((((.(.(((....((-((((.((.-....))))))))....((((......))))))).))))).......))). ( -25.30, z-score = -1.37, R) >droPer1.super_24 1044420 86 - 1556852 UCAUCGUCAU-UAUCGUUUUGCCAAAAUCCAAAUAAAAUCAACAAAAAAAAGCCAAAUAAACAGGUUCCCUUAAUGGCCAUUAGGCA--------------------- ..........-........((((...........................((((.........)))).((.....))......))))--------------------- ( -7.60, z-score = 0.29, R) >consensus UCCACGCCAUGUGCCGCAUGGAGAUAAUC_AUUCGAAACCGAAAAAAAAGAACCCCA_AAACGGGUUCUUCUAAUAGCCAUUAAGCUAUCGUCUCACUCUCAAAUACG ((((.((........)).))))...........................((((((.......)))))).....(((((......)))))................... (-11.71 = -12.10 + 0.39)


| Location | 4,442,258 – 4,442,363 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 78.55 |
| Shannon entropy | 0.41131 |
| G+C content | 0.43343 |
| Mean single sequence MFE | -33.90 |
| Consensus MFE | -19.66 |
| Energy contribution | -22.55 |
| Covariance contribution | 2.89 |
| Combinations/Pair | 1.28 |
| Mean z-score | -3.04 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.95 |
| SVM RNA-class probability | 0.996551 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
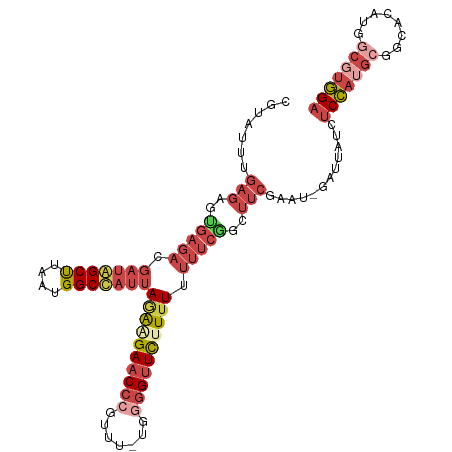
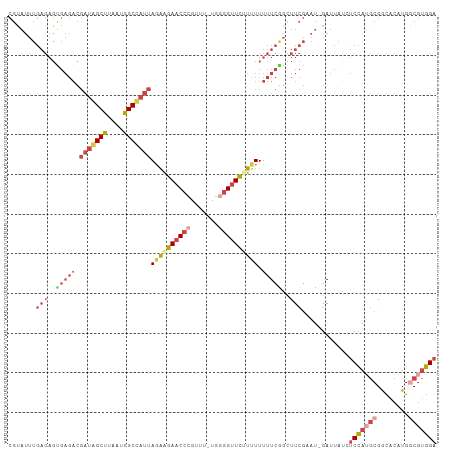
>dm3.chr3L 4442258 105 - 24543557 CGUAUUUGAGAGUGAGACGAUAGCUUAAUGGCUAUUAGAAGAACCCGUUU-UGCGGUUC-UUUUUUUCGGCUUCGAAU-GAUUAUUUCCAUGCGGCACAUGGCGUGGA ..((((((((..(((((.(((((((....)))))))(((((((((.....-...)))))-)))))))))..)))))))-)......(((((((........))))))) ( -34.10, z-score = -2.79, R) >droSim1.chr3L 3977802 106 - 22553184 CGCAUUUGAGAGUGAGACGAUAGCUUAAUGGCUAUUAGAAGAACCCGUUU-UGGGGUUCUUUUUUUUCGGUUUCGAAU-GAUUAUCUCCAUGCGGCACAUGGCGUGGA ..(((((((((.(((((.(((((((....)))))))((((((((((....-..)))))))))).))))).))))))))-)......(((((((........))))))) ( -43.70, z-score = -5.60, R) >droSec1.super_2 4431041 106 - 7591821 CGCAUUUGAGAGUGAGACGAUAGCUUAAUGGCUAUUAGAAGAACCCGUUU-UGGGGUUCUUUUUUUUCGGUUUCGAAU-GAUUAUCUCCAUGCGGCACAUGGCGUGGA ..(((((((((.(((((.(((((((....)))))))((((((((((....-..)))))))))).))))).))))))))-)......(((((((........))))))) ( -43.70, z-score = -5.60, R) >droYak2.chr3L 5019729 106 - 24197627 CGUAUUUGAGAGUGAGACGAUGGCUUAAUGGCCAUUAGAAGAACCCGUUU-UGGGGUUCUUAUUUUUCAGCUUCGUAU-GAUUAUCUCCAGGCGGCACAUGGCGUGGA (((((..(((..(((((.(((((((....)))))))..((((((((....-..))))))))...))))).))).))))-)......((((.((........)).)))) ( -35.40, z-score = -2.54, R) >droEre2.scaffold_4784 7150872 105 - 25762168 CGUAUUUGAGAGCGAGACGAUGGCUUAAUGGCCAUUAGAAGAACCCGUUU-UGGUGUUC-UUUUUUUCGGCUUCGCAU-GAUUACCUCCAUGCGGCACAUGACGUGGA (((...((.((((((((.(((((((....)))))))((((((((((....-.)).))))-)))))))).))))(((((-(........))))))...))..))).... ( -33.50, z-score = -2.16, R) >droPer1.super_24 1044420 86 + 1556852 ---------------------UGCCUAAUGGCCAUUAAGGGAACCUGUUUAUUUGGCUUUUUUUUGUUGAUUUUAUUUGGAUUUUGGCAAAACGAUA-AUGACGAUGA ---------------------.(((....)))((((((((...)))...............((((((..(..((.....))..)..))))))...))-)))....... ( -13.00, z-score = 0.41, R) >consensus CGUAUUUGAGAGUGAGACGAUAGCUUAAUGGCCAUUAGAAGAACCCGUUU_UGGGGUUCUUUUUUUUCGGCUUCGAAU_GAUUAUCUCCAUGCGGCACAUGGCGUGGA .......(((..(((((.(((((((....)))))))((((((((((.......)))))))))).)))))..)))............(((((((........))))))) (-19.66 = -22.55 + 2.89)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:02:47 2011