| Sequence ID | dm3.chr3L |
|---|---|
| Location | 4,424,089 – 4,424,186 |
| Length | 97 |
| Max. P | 0.578294 |

| Location | 4,424,089 – 4,424,186 |
|---|---|
| Length | 97 |
| Sequences | 8 |
| Columns | 110 |
| Reading direction | forward |
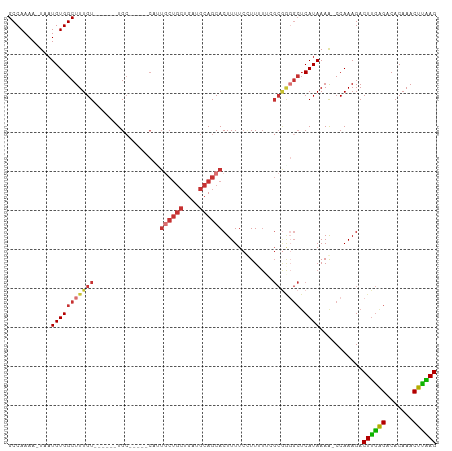
| Mean pairwise identity | 81.85 |
| Shannon entropy | 0.35056 |
| G+C content | 0.44202 |
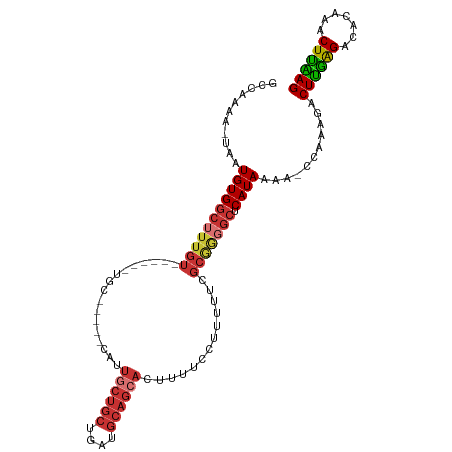
| Mean single sequence MFE | -27.76 |
| Consensus MFE | -16.98 |
| Energy contribution | -17.70 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.578294 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
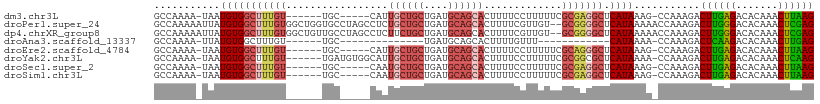
>dm3.chr3L 4424089 97 + 24543557 GCCAAAA-UAAUGUGGCUUUGU------UGC-----CAUUGCUGCUGAUGCAGCACUUUUCCUUUUUCGCGAGGCUCAUAAAG-CCAAAGACUUGAGACACAAACUUAAG .......-.....(((((((((------.((-----(..((((((....))))))....(((......).)))))..))))))-)))....((((((.......)))))) ( -28.30, z-score = -2.29, R) >droPer1.super_24 1023995 108 - 1556852 GCCAAAAAUUAUGUGGCUUUGUGGCUGGUGCCUAGCCUCUGCUGCUGAUGCAGCACUUUUCGUUGU--GCGGGGCUCAUAAAAACCAAAGACUUGGGACACAAACUCGAG ........(((((.((((((((((((((...))))))..((((((....))))))...........--)))))))))))))..........((((((.......)))))) ( -33.10, z-score = -0.77, R) >dp4.chrXR_group8 2516944 108 + 9212921 GCCAAAAAUUAUGUGGCUUUGUGGCUGUUGCCUAGCCUCUUCUGCUGAUGCAGCACUUUUCGUUGU--GCGGGGCUCAUAAAAACCAAAGACUUGGGACACAAACUCGAG ((((.........))))((((((((....))).((((((...(((....)))((((........))--))))))))........((((....))))...)))))...... ( -30.30, z-score = -0.64, R) >droAna3.scaffold_13337 2505391 76 + 23293914 GCCAAAA-UUAUGUGGCUUUGU------UGC--------------UGAUGCAGCACUUUUGUUU------------CAUAAAA-CCAAAGACUCAAGACACAAACUUGAG ((((...-.....))))..(((------(((--------------....))))))((((.((((------------....)))-).)))).((((((.......)))))) ( -20.30, z-score = -2.58, R) >droEre2.scaffold_4784 7132950 97 + 25762168 GCCAAAA-UAAUGUGGCUUUGU------UGC-----CAUUGCUGCUGAUGCAGCACUUUUCCUUUUUCGCAGGGCUCAUAAAG-CCAAAGACUUGAGACACAAACUUAAG .......-.....(((((((((------.((-----(..((((((....))))))......((.......)))))..))))))-)))....((((((.......)))))) ( -27.60, z-score = -1.70, R) >droYak2.chr3L 5000511 102 + 24197627 GCCAAAA-UAAUGUGGCUUUGU------UGAUGUGGCAUUGCUGCUGAUGCAGCACUUUUCCUUUUUCGCGGCGCUCAUAAAA-CCAAAGACUUGAGACACAAACUCAAG (((....-.(((((.((.....------....)).)))))(((((....)))))................)))..........-.......((((((.......)))))) ( -25.70, z-score = -0.69, R) >droSec1.super_2 4413176 97 + 7591821 GCCAAAA-UAAUGUGGCUUUGU------UGC-----CAAUGCUGCUGAUGCAGCACUUUUCCUUUUUCGCGAGGCUCAUAAAG-CCAAAGACUUGAGACACAAACUUAAG .......-.....(((((((((------.((-----(..((((((....))))))....(((......).)))))..))))))-)))....((((((.......)))))) ( -28.40, z-score = -2.33, R) >droSim1.chr3L 3960816 97 + 22553184 GCCAAAA-UAAUGUGGCUUUGU------UGC-----CAAUGCUGCUGAUGCAGCACUUUUCCUUUUUCGCGAGGCUCAUAAAG-CCAAAGACUUGAGACACAAACUUAAG .......-.....(((((((((------.((-----(..((((((....))))))....(((......).)))))..))))))-)))....((((((.......)))))) ( -28.40, z-score = -2.33, R) >consensus GCCAAAA_UAAUGUGGCUUUGU______UGC_____CAUUGCUGCUGAUGCAGCACUUUUCCUUUUUCGCGGGGCUCAUAAAA_CCAAAGACUUGAGACACAAACUUAAG ...........(((((((((((.................((((((....)))))).............))))))).))))...........((((((.......)))))) (-16.98 = -17.70 + 0.72)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:02:44 2011