
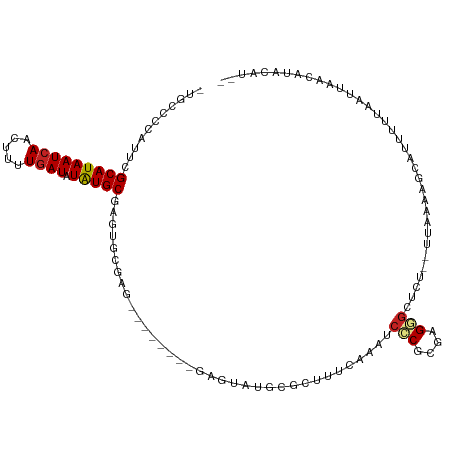
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 4,312,724 – 4,312,826 |
| Length | 102 |
| Max. P | 0.886542 |

| Location | 4,312,724 – 4,312,826 |
|---|---|
| Length | 102 |
| Sequences | 11 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 73.07 |
| Shannon entropy | 0.55393 |
| G+C content | 0.36920 |
| Mean single sequence MFE | -22.98 |
| Consensus MFE | -8.63 |
| Energy contribution | -8.75 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.886542 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2L 4312724 102 + 23011544 -UGCCCCAUUCGCAUAAUCAACUUUUGAUAUAUGCGAGUGCGAG--------GAGUAUGCGCUUUCAAAUCCCGCGAGGGCUCU--UUAAAAGCAUUUUUAAUUAACAUACAU-- -..(((((((((((((((((.....)))).)))))))))).).)--------).(((((.(((((.(((.(((....)))...)--)).)))))..((......)))))))..-- ( -30.90, z-score = -3.53, R) >droSim1.chr2L 4235031 102 + 22036055 -UGCCCCAUUCGCAUAAUCAACUUUUGAUAUAUGCGAGUGCGAG--------GAGUAUGCGCUUUCAAAUCCCGCGAGGG--CUCUUUAAAAGCAUUUUUAAUUAACAUACAU-- -..(((((((((((((((((.....)))).)))))))))).).)--------).(((((.(((((.(((.(((....)))--...))).)))))..((......)))))))..-- ( -30.90, z-score = -3.53, R) >droSec1.super_5 2410432 102 + 5866729 -UGCCCCAUUCGCAUAAUCAACUUUUGAUAUAUGCGAGUGCGAG--------GAGUAUGCGCUUUCAAAUCCCGCGAGGGCUCU--UUAAAUGCAUUUUUAAUUAACAUACAU-- -..(((((((((((((((((.....)))).)))))))))).).)--------).(((((...........(((....)))....--(((((......)))))....)))))..-- ( -28.90, z-score = -2.75, R) >droYak2.chr2L 4343732 102 + 22324452 -UGCCCCAUUCGCAUAAUCAACUUUUGAUAUAUGCGAGUGCGAG--------GAGUAUGCGCUUUCAAAUCCCGCGAGGGCUCU--UUAAAAGCAUUUUUAAUUAACAUACAU-- -..(((((((((((((((((.....)))).)))))))))).).)--------).(((((.(((((.(((.(((....)))...)--)).)))))..((......)))))))..-- ( -30.90, z-score = -3.53, R) >droEre2.scaffold_4929 4389594 102 + 26641161 -UGCCCCAUUCGCAUAAUCAACUUUUGAUAUAUGCGAGUGCGAG--------GAGUAUGCGCUUUCAAAUCCCGCGAGGGCUCU--UUAAAAGCAUUUUUAAUUAACAUACAU-- -..(((((((((((((((((.....)))).)))))))))).).)--------).(((((.(((((.(((.(((....)))...)--)).)))))..((......)))))))..-- ( -30.90, z-score = -3.53, R) >droAna3.scaffold_12916 1510212 102 + 16180835 -GGGGCUAUUCGCAUAAUCAAGUUUUAAUAUAUGCCAGCGAAUGAGU------GGAUUGCGCUUUAAAGUCCCCAGGGU----UGCUAAAUAAGUUUUUUUCCAAAAACAUUU-- -((((((((((((........................))))))((((------(.....)))))...))))))..(((.----.(((.....))).....)))..........-- ( -19.46, z-score = 0.48, R) >dp4.chr4_group2 307571 113 + 1235136 UGCCGCCAUUCGCAUAAUCAACUUUUGAUAUAUGCAAGACAGAGUGUAGAGUGUAUAUGCGCUUUCAAAUCUCCCGAGAGAGCUCUUUAAAAGCAUUUUUAAUUAGAAUACAU-- .......(((((((((((((.....)))).))))).....((((((.(((((((....))))))))...((((....)))))))))...................))))....-- ( -22.90, z-score = -1.02, R) >droPer1.super_8 384201 113 - 3966273 UGCCGCCAUUCGCAUAAUCAACUUUUGAUAUAUGCAAGACAGAGUGUAGAGUGUAUAUGCGCUUUCAAAUCUCCCGAGAGAGCUCUUUAAAAGCAUUUUUAAUUAGAAUACAU-- .......(((((((((((((.....)))).))))).....((((((.(((((((....))))))))...((((....)))))))))...................))))....-- ( -22.90, z-score = -1.02, R) >droWil1.scaffold_180772 2036743 84 + 8906247 ----GCCCCUUGCAUAAUCAACUUUUGAUAUGUGC--------------------UAUGCA-----AAAUCUCCCGAGAG--CUCUUUAAAAGCAUUUUUAAUUAACAUAUAUCU ----((.....)).............((((((((.--------------------......-----...(((....)))(--((.......)))............)))))))). ( -10.60, z-score = -0.64, R) >droVir3.scaffold_13246 383900 85 + 2672878 -UGUCCUGUCCGCAUAAUCAACUUUUGAUAUAUGCG-------------------------CUUUCAAAUCUCUUGUGUGCUCU--UUAAAAACAUUUCUAAUUAACAUAUAU-- -..........(((((((((.....)))).)))))(-------------------------(...(((.....)))...))...--...........................-- ( -9.50, z-score = -1.31, R) >droMoj3.scaffold_6500 384165 86 + 32352404 --GUUCUAUGCGCAUAAUCAACUUUUGAUAUAUGCG-------------------------CUUUUAAAUCUCUCGUGUGCUCUCUUUAAAAGCAUCUCUAAUUAACAUAUAU-- --.......(((((((((((.....)))).))))))-------------------------)...............(((((.........))))).................-- ( -14.90, z-score = -2.88, R) >consensus _UGCCCCAUUCGCAUAAUCAACUUUUGAUAUAUGCGAGUGCGAG________GAGUAUGCGCUUUCAAAUCCCGCGAGGGCUCU__UUAAAAGCAUUUUUAAUUAACAUACAU__ ...........(((((((((.....)))).)))))...................................(((....)))................................... ( -8.63 = -8.75 + 0.13)

| Location | 4,312,724 – 4,312,826 |
|---|---|
| Length | 102 |
| Sequences | 11 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 73.07 |
| Shannon entropy | 0.55393 |
| G+C content | 0.36920 |
| Mean single sequence MFE | -25.54 |
| Consensus MFE | -8.54 |
| Energy contribution | -8.74 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.737891 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
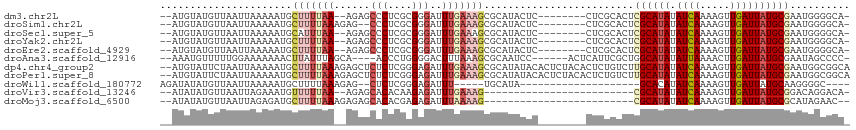
>dm3.chr2L 4312724 102 - 23011544 --AUGUAUGUUAAUUAAAAAUGCUUUUAA--AGAGCCCUCGCGGGAUUUGAAAGCGCAUACUC--------CUCGCACUCGCAUAUAUCAAAAGUUGAUUAUGCGAAUGGGGCA- --..((((((...........((((((((--(...(((....))).))))))))))))))).(--------((((...(((((((.((((.....))))))))))).)))))..- ( -32.30, z-score = -3.05, R) >droSim1.chr2L 4235031 102 - 22036055 --AUGUAUGUUAAUUAAAAAUGCUUUUAAAGAG--CCCUCGCGGGAUUUGAAAGCGCAUACUC--------CUCGCACUCGCAUAUAUCAAAAGUUGAUUAUGCGAAUGGGGCA- --..((((((...........(((((((((...--(((....))).))))))))))))))).(--------((((...(((((((.((((.....))))))))))).)))))..- ( -32.30, z-score = -3.05, R) >droSec1.super_5 2410432 102 - 5866729 --AUGUAUGUUAAUUAAAAAUGCAUUUAA--AGAGCCCUCGCGGGAUUUGAAAGCGCAUACUC--------CUCGCACUCGCAUAUAUCAAAAGUUGAUUAUGCGAAUGGGGCA- --((((((...........))))))....--...(((((.((((((..((......))...))--------).)))..(((((((.((((.....)))))))))))..))))).- ( -29.90, z-score = -2.27, R) >droYak2.chr2L 4343732 102 - 22324452 --AUGUAUGUUAAUUAAAAAUGCUUUUAA--AGAGCCCUCGCGGGAUUUGAAAGCGCAUACUC--------CUCGCACUCGCAUAUAUCAAAAGUUGAUUAUGCGAAUGGGGCA- --..((((((...........((((((((--(...(((....))).))))))))))))))).(--------((((...(((((((.((((.....))))))))))).)))))..- ( -32.30, z-score = -3.05, R) >droEre2.scaffold_4929 4389594 102 - 26641161 --AUGUAUGUUAAUUAAAAAUGCUUUUAA--AGAGCCCUCGCGGGAUUUGAAAGCGCAUACUC--------CUCGCACUCGCAUAUAUCAAAAGUUGAUUAUGCGAAUGGGGCA- --..((((((...........((((((((--(...(((....))).))))))))))))))).(--------((((...(((((((.((((.....))))))))))).)))))..- ( -32.30, z-score = -3.05, R) >droAna3.scaffold_12916 1510212 102 - 16180835 --AAAUGUUUUUGGAAAAAAACUUAUUUAGCA----ACCCUGGGGACUUUAAAGCGCAAUCC------ACUCAUUCGCUGGCAUAUAUUAAAACUUGAUUAUGCGAAUAGCCCC- --....((((((....))))))..........----.....((((.((....((((.(((..------....))))))).(((((.((((.....)))))))))....))))))- ( -19.60, z-score = -0.61, R) >dp4.chr4_group2 307571 113 - 1235136 --AUGUAUUCUAAUUAAAAAUGCUUUUAAAGAGCUCUCUCGGGAGAUUUGAAAGCGCAUAUACACUCUACACUCUGUCUUGCAUAUAUCAAAAGUUGAUUAUGCGAAUGGCGGCA --.(((((............((((((((((...((((....)))).))))))))))...))))).........((((((((((((.((((.....)))))))))))..))))).. ( -27.36, z-score = -1.83, R) >droPer1.super_8 384201 113 + 3966273 --AUGUAUUCUAAUUAAAAAUGCUUUUAAAGAGCUCUCUCGGGAGAUUUGAAAGCGCAUAUACACUCUACACUCUGUCUUGCAUAUAUCAAAAGUUGAUUAUGCGAAUGGCGGCA --.(((((............((((((((((...((((....)))).))))))))))...))))).........((((((((((((.((((.....)))))))))))..))))).. ( -27.36, z-score = -1.83, R) >droWil1.scaffold_180772 2036743 84 - 8906247 AGAUAUAUGUUAAUUAAAAAUGCUUUUAAAGAG--CUCUCGGGAGAUUU-----UGCAUA--------------------GCACAUAUCAAAAGUUGAUUAUGCAAGGGGC---- .(((((.(((((..(((((..((((.....)))--)..((....)))))-----))..))--------------------))).)))))......................---- ( -12.90, z-score = 0.39, R) >droVir3.scaffold_13246 383900 85 - 2672878 --AUAUAUGUUAAUUAGAAAUGUUUUUAA--AGAGCACACAAGAGAUUUGAAAG-------------------------CGCAUAUAUCAAAAGUUGAUUAUGCGGACAGGACA- --.....((((..........((((((((--(...(......)...))))))))-------------------------)(((((.((((.....))))))))).)))).....- ( -15.80, z-score = -2.18, R) >droMoj3.scaffold_6500 384165 86 - 32352404 --AUAUAUGUUAAUUAGAGAUGCUUUUAAAGAGAGCACACGAGAGAUUUAAAAG-------------------------CGCAUAUAUCAAAAGUUGAUUAUGCGCAUAGAAC-- --...........(((((..(((((((...)))))))..(....).)))))..(-------------------------((((((.((((.....))))))))))).......-- ( -18.80, z-score = -3.00, R) >consensus __AUGUAUGUUAAUUAAAAAUGCUUUUAA__AGAGCCCUCGCGGGAUUUGAAAGCGCAUACUC________CUCGCACUCGCAUAUAUCAAAAGUUGAUUAUGCGAAUGGGGCA_ ...................................(((....)))..................................((((((.((((.....)))))))))).......... ( -8.54 = -8.74 + 0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:16:02 2011