
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 4,403,068 – 4,403,166 |
| Length | 98 |
| Max. P | 0.799697 |

| Location | 4,403,068 – 4,403,166 |
|---|---|
| Length | 98 |
| Sequences | 9 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 73.31 |
| Shannon entropy | 0.52724 |
| G+C content | 0.35773 |
| Mean single sequence MFE | -25.57 |
| Consensus MFE | -8.09 |
| Energy contribution | -8.74 |
| Covariance contribution | 0.65 |
| Combinations/Pair | 1.54 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.799697 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
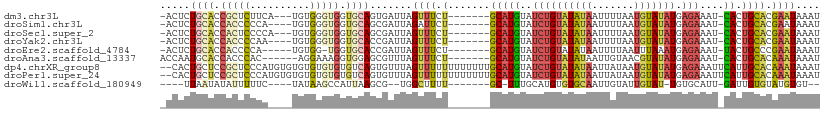
>dm3.chr3L 4403068 98 + 24543557 -ACUCUGCACCGCUCUUCA---UGUGGGUGGUGCAGUGAUUAGUUUCU-------GCAUGUAUCUGUAUAUAAUUUUAAUGUAUAUGAGAAAU-CACUGCACGAAUAAAU -.(.(((((((((((....---...))))))))))).)....((((.(-------(((((..((((((((((.......))))))).)))...-)).)))).)))).... ( -31.40, z-score = -3.75, R) >droSim1.chr3L 3939602 97 + 22553184 -ACUCUGCACCACCCCCA----UGUGGGUGGUGCAGCGAUUAGAUUCU-------GCAUGUAUCUGUAUAUAAUUUUAAUGUAUAUGAGAAAU-CACUGCACGAAUAAAU -.(.(((((((((((...----...))))))))))).).....(((((-------(((((..((((((((((.......))))))).)))...-)).)))).)))).... ( -34.70, z-score = -4.89, R) >droSec1.super_2 4392556 98 + 7591821 -ACUCUGCACCACUCCCCA---UGUGGGUGGUGCAGCGAUUAGUUUCU-------GCAUGUAUCUGUAUAUAAUUUUAAUGUAUAUGAGAAAU-CACUGCACGAAUAAAU -.(.(((((((((((....---...))))))))))).)....((((.(-------(((((..((((((((((.......))))))).)))...-)).)))).)))).... ( -30.30, z-score = -3.35, R) >droYak2.chr3L 4979078 97 + 24197627 -ACUCUGCACCACCCCAA----UGUGGGUGGUGCACCGAUUAGUUUCU-------GCAUGUAUCUGUAUAUAAUUUUAAUGUAUAUGAGAAAU-CACUGCACGAAUAAAU -....((((((((((...----...)))))))))).......((((.(-------(((((..((((((((((.......))))))).)))...-)).)))).)))).... ( -30.20, z-score = -3.73, R) >droEre2.scaffold_4784 7111637 95 + 25762168 -ACUCUGCACCACCCCA-----UGUGG-UGGUGCACCGAUUAGUUUCU-------GCAUGUAUCUGUAUAUAAUUUUAAUUUAAAUGAGAAAU-UACUGCCCGAAUAAAU -....(((((((((...-----...))-))))))).....((((((((-------.(((.......((........))......)))))))))-)).............. ( -22.12, z-score = -2.56, R) >droAna3.scaffold_13337 2483306 96 + 23293914 ACCAAUGCACCACCCAC------AGGAAAGGUGGAGCGUUUAGUUUCU-------GCAUGUAUCUGUAUAUAAUUGUAACGUAUAUGAGAAAU-CACUGCACAAAUAAAU ...(((((.(((((...------......))))).)))))..((((((-------.(((((((...((((....))))..)))))))))))))-................ ( -23.20, z-score = -1.74, R) >dp4.chrXR_group8 2495376 108 + 9212921 --CACUGCUCCGCUCCCAUGUGUGUGUGUGUGUCAGUGUUUAGUUUUUUUUUUUUGCAUGUAUCUGUAUAUAAUUAUAAUGUAUAUGAGAAAUUCAUUGCACAAAUAAAU --(((.((..(((......))).)))))((((.(((((............(((((.(((((((...((((....))))..))))))))))))..)))))))))....... ( -18.54, z-score = -0.26, R) >droPer1.super_24 1001603 108 - 1556852 --CACUGCUCCGCUCCCAUGUGUGUGUGUGUGUCAGUGUUUAGUUUUUUUUUUUUGCAUGUAUCUGUAUAUAAUUAUAAUGUAUAUGAGAAAUUCAUUGCACAAAUAAAU --(((.((..(((......))).)))))((((.(((((............(((((.(((((((...((((....))))..))))))))))))..)))))))))....... ( -18.54, z-score = -0.26, R) >droWil1.scaffold_180949 1053709 88 + 6375548 ----UUAAUAUAUUUUUC----UAUAAGCCAUUAAGCG--UGGCUUUU-------GC-UUUGCAUGUGUGCAAUUGUAUUGUAU-UGUGCAUU-GAUUGUGUAUGUGU-- ----..............----...(((((((.....)--))))))..-------..-...(((((((..(((((((((.....-.)))))..-.))))..)))))))-- ( -21.10, z-score = -1.15, R) >consensus _ACUCUGCACCACUCCCA____UGUGGGUGGUGCAGCGAUUAGUUUCU_______GCAUGUAUCUGUAUAUAAUUUUAAUGUAUAUGAGAAAU_CACUGCACGAAUAAAU .....((((.(((((..........))))).))))....................(((((..((((((((((.......))))))).)))....)).))).......... ( -8.09 = -8.74 + 0.65)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:02:41 2011