
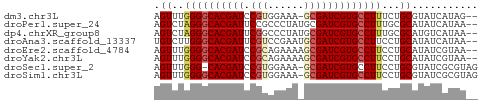
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 4,385,784 – 4,385,835 |
| Length | 51 |
| Max. P | 0.998612 |

| Location | 4,385,784 – 4,385,835 |
|---|---|
| Length | 51 |
| Sequences | 8 |
| Columns | 54 |
| Reading direction | forward |
| Mean pairwise identity | 78.63 |
| Shannon entropy | 0.41008 |
| G+C content | 0.53592 |
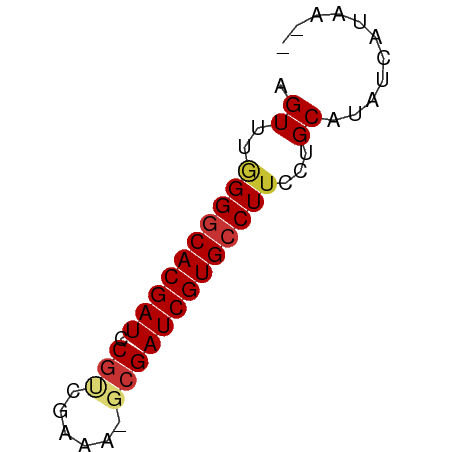
| Mean single sequence MFE | -20.58 |
| Consensus MFE | -17.41 |
| Energy contribution | -17.75 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.13 |
| Mean z-score | -3.01 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.42 |
| SVM RNA-class probability | 0.998612 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
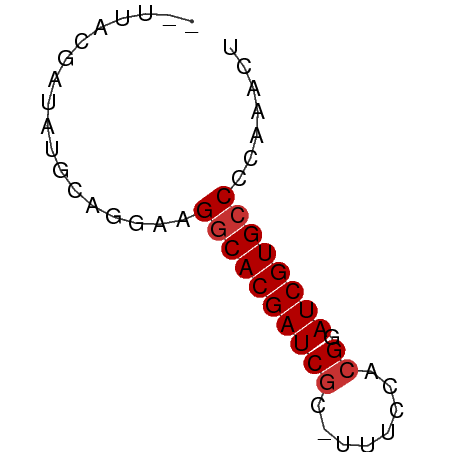
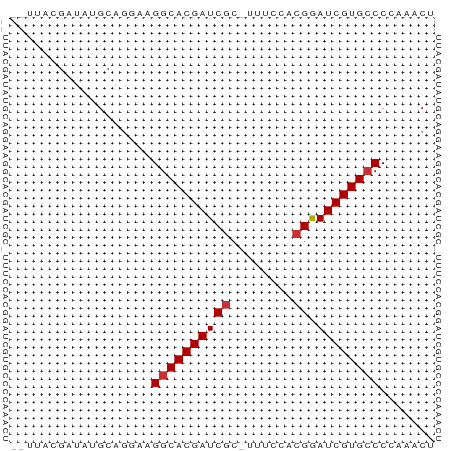
>dm3.chr3L 4385784 51 + 24543557 AGUUUGGGGCACGAUCCGUGGAAA-GCGAUCGUGCCUUUCUGCGUAUCAUAG-- .((..(((((((((((.((.....-)))))))))))))...)).........-- ( -20.30, z-score = -3.11, R) >droPer1.super_24 981463 52 - 1556852 AGUCUAGGGCACGAUUCCGCCCUAUGCGAUCGUGCCUUUGCGCAUAUCAUAA-- .((..((((((((((..(((.....))))))))))))).))...........-- ( -18.90, z-score = -2.91, R) >dp4.chrXR_group8 2475175 52 + 9212921 AGUCUAGGGCACGAUUCGGCCCUAUGCGAUCGUGCCUUUGCGCAUGUCAUAA-- .((.((((((.((...)))))))).))((.(((((......)))))))....-- ( -18.90, z-score = -2.04, R) >droAna3.scaffold_13337 2465081 52 + 23293914 UGUCUUGGGCACGAUUCGUCCGAAUGCGAUCGUGCCUUCCUGCAUAUCAUAA-- (((...(((((((((.(((......))))))))))))....)))........-- ( -18.40, z-score = -3.11, R) >droEre2.scaffold_4784 7093486 52 + 25762168 AGUUUGGGGCACGAUCCGCAGAAAAGCGAUCGUGCCUUCCUGCAUAUCGUAA-- .((..(((((((((((.((......)))))))))))))...)).........-- ( -21.40, z-score = -3.43, R) >droYak2.chr3L 4961405 52 + 24197627 AGUUUGGGGCACGAUCCGCAGAAAAGCGAUCGUGCCUUCCUGCAUAUCGUAA-- .((..(((((((((((.((......)))))))))))))...)).........-- ( -21.40, z-score = -3.43, R) >droSec1.super_2 4375598 52 + 7591821 AGUUUGGG-CACGAUCCGUGGAAA-GCGAUCGUGCCUUCCUGCGUAUCGCGUAG .....(((-(((((((.((.....-))))))))))))..(((((.....))))) ( -22.40, z-score = -3.14, R) >droSim1.chr3L 3922135 53 + 22553184 AGUUUGGGGCACGAUCCGUGGAAA-GCGAUCGUGCCUUCCUGCGUAUCGCGUAG .....(((((((((((.((.....-))))))))))))).(((((.....))))) ( -22.90, z-score = -2.89, R) >consensus AGUUUGGGGCACGAUCCGUCGAAA_GCGAUCGUGCCUUCCUGCAUAUCAUAA__ .((..((((((((((.(((......)))))))))))))...))........... (-17.41 = -17.75 + 0.34)
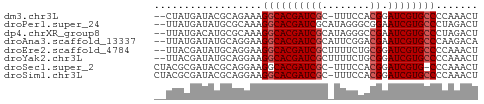
| Location | 4,385,784 – 4,385,835 |
|---|---|
| Length | 51 |
| Sequences | 8 |
| Columns | 54 |
| Reading direction | reverse |
| Mean pairwise identity | 78.63 |
| Shannon entropy | 0.41008 |
| G+C content | 0.53592 |
| Mean single sequence MFE | -17.86 |
| Consensus MFE | -13.15 |
| Energy contribution | -13.40 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.04 |
| SVM RNA-class probability | 0.997134 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 4385784 51 - 24543557 --CUAUGAUACGCAGAAAGGCACGAUCGC-UUUCCACGGAUCGUGCCCCAAACU --................(((((((((..-........)))))))))....... ( -15.40, z-score = -2.79, R) >droPer1.super_24 981463 52 + 1556852 --UUAUGAUAUGCGCAAAGGCACGAUCGCAUAGGGCGGAAUCGUGCCCUAGACU --................(((((((((((.....)))..))))))))....... ( -16.20, z-score = -1.52, R) >dp4.chrXR_group8 2475175 52 - 9212921 --UUAUGACAUGCGCAAAGGCACGAUCGCAUAGGGCCGAAUCGUGCCCUAGACU --................(((((((((((.....).)).))))))))....... ( -14.30, z-score = -0.91, R) >droAna3.scaffold_13337 2465081 52 - 23293914 --UUAUGAUAUGCAGGAAGGCACGAUCGCAUUCGGACGAAUCGUGCCCAAGACA --................((((((((((........)).))))))))....... ( -14.40, z-score = -1.66, R) >droEre2.scaffold_4784 7093486 52 - 25762168 --UUACGAUAUGCAGGAAGGCACGAUCGCUUUUCUGCGGAUCGUGCCCCAAACU --.((((((.(((((((((((......))))))))))).))))))......... ( -22.50, z-score = -4.42, R) >droYak2.chr3L 4961405 52 - 24197627 --UUACGAUAUGCAGGAAGGCACGAUCGCUUUUCUGCGGAUCGUGCCCCAAACU --.((((((.(((((((((((......))))))))))).))))))......... ( -22.50, z-score = -4.42, R) >droSec1.super_2 4375598 52 - 7591821 CUACGCGAUACGCAGGAAGGCACGAUCGC-UUUCCACGGAUCGUG-CCCAAACU ...((((((.((..(((((((......))-))))).)).))))))-........ ( -18.70, z-score = -2.98, R) >droSim1.chr3L 3922135 53 - 22553184 CUACGCGAUACGCAGGAAGGCACGAUCGC-UUUCCACGGAUCGUGCCCCAAACU ....(((...))).((..(((((((((..-........)))))))))))..... ( -18.90, z-score = -2.94, R) >consensus __UUACGAUAUGCAGGAAGGCACGAUCGC_UUUCCACGGAUCGUGCCCCAAACU ..................((((((((((........)).))))))))....... (-13.15 = -13.40 + 0.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:02:39 2011