
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 4,343,294 – 4,343,385 |
| Length | 91 |
| Max. P | 0.701980 |

| Location | 4,343,294 – 4,343,385 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 81.38 |
| Shannon entropy | 0.32740 |
| G+C content | 0.49991 |
| Mean single sequence MFE | -20.24 |
| Consensus MFE | -16.38 |
| Energy contribution | -16.22 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.701980 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
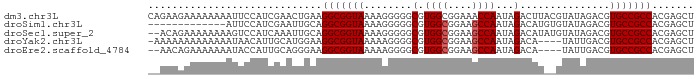
>dm3.chr3L 4343294 91 + 24543557 AGCUCGUGGCGGCACGUCUAUACGUAAGUCUAUUGGUUUCCGCCACGCCCCCUUUUACCGCCUUCAGUUCGAUGGAAUUUUUUUUCUUCUG ....((((((((.....(((.((....))....)))...)))))))).......................((.((((.....)))).)).. ( -18.90, z-score = -0.54, R) >droSim1.chr3L 3880629 78 + 22553184 AGCUCGUGGCGGCACGUCUAUACACAUGUCUAUUGGCUUCCGCCACGCCCCCUUUUACCGCCUGCAAUUCGAUGGAAU------------- ....((((((((...(((.(((........))).)))..))))))))..........(((.(........).)))...------------- ( -18.20, z-score = 0.15, R) >droSec1.super_2 4335604 89 + 7591821 AGCUCGUGGCGGCACGUCUAUACAUAUGUCUAUUGGCUUCCGCCACGCCCCCUUUUACCGCCUGCAAUUUGAUGGACUUUUUUUUCUGU-- ....((((((((...(((.(((........))).)))..))))))))..........(((.(........).)))..............-- ( -18.80, z-score = -0.27, R) >droYak2.chr3L 4918647 86 + 24197627 AGCUCGUGGCGGCACGUCAAUA----UGUCUAUUGGCUUCCGCCACGCCCCUUUUUACCGCCUUCCAUGCAAUGUUAUUUUUUUUUUUUU- ....((((((((...(((((((----....)))))))..))))))))............((.......))....................- ( -21.70, z-score = -2.86, R) >droEre2.scaffold_4784 7052704 85 + 25762168 AGCUCGUGGCGGCACGUCAAUA----UGUCUAUUGGCUUCCGCCACGCCCCUUUUUACCGCCUUCCCUGCAAUGGUAUUUUUUUCUGUU-- ....((((((((...(((((((----....)))))))..))))))))........((((((.......))...))))............-- ( -23.60, z-score = -2.52, R) >consensus AGCUCGUGGCGGCACGUCUAUAC__AUGUCUAUUGGCUUCCGCCACGCCCCCUUUUACCGCCUUCAAUUCGAUGGAAUUUUUUUUCUUU__ ....((((((((...(((.(((........))).)))..))))))))............................................ (-16.38 = -16.22 + -0.16)


| Location | 4,343,294 – 4,343,385 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 81.38 |
| Shannon entropy | 0.32740 |
| G+C content | 0.49991 |
| Mean single sequence MFE | -23.53 |
| Consensus MFE | -18.43 |
| Energy contribution | -18.63 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.571868 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 4343294 91 - 24543557 CAGAAGAAAAAAAAUUCCAUCGAACUGAAGGCGGUAAAAGGGGGCGUGGCGGAAACCAAUAGACUUACGUAUAGACGUGCCGCCACGAGCU ..............((((.....((((....)))).....))))((((((((.............(((((....))))))))))))).... ( -22.41, z-score = -0.82, R) >droSim1.chr3L 3880629 78 - 22553184 -------------AUUCCAUCGAAUUGCAGGCGGUAAAAGGGGGCGUGGCGGAAGCCAAUAGACAUGUGUAUAGACGUGCCGCCACGAGCU -------------......(((..((((.....))))..((.(((.((((....))))......((((......))))))).)).)))... ( -23.60, z-score = -0.26, R) >droSec1.super_2 4335604 89 - 7591821 --ACAGAAAAAAAAGUCCAUCAAAUUGCAGGCGGUAAAAGGGGGCGUGGCGGAAGCCAAUAGACAUAUGUAUAGACGUGCCGCCACGAGCU --...........................(((((((.......((.((((....))))(((....))))).......)))))))....... ( -22.64, z-score = -0.63, R) >droYak2.chr3L 4918647 86 - 24197627 -AAAAAAAAAAAAAUAACAUUGCAUGGAAGGCGGUAAAAAGGGGCGUGGCGGAAGCCAAUAGACA----UAUUGACGUGCCGCCACGAGCU -.................(((((.......))))).....((..((((((((..(.(((((....----))))).)...))))))))..)) ( -24.30, z-score = -2.30, R) >droEre2.scaffold_4784 7052704 85 - 25762168 --AACAGAAAAAAAUACCAUUGCAGGGAAGGCGGUAAAAAGGGGCGUGGCGGAAGCCAAUAGACA----UAUUGACGUGCCGCCACGAGCU --............((((...((.......))))))....((..((((((((..(.(((((....----))))).)...))))))))..)) ( -24.70, z-score = -1.76, R) >consensus __AAAAAAAAAAAAUUCCAUCGAAUUGAAGGCGGUAAAAGGGGGCGUGGCGGAAGCCAAUAGACAU__GUAUAGACGUGCCGCCACGAGCU .............................(((((((..........((((....))))...................)))))))....... (-18.43 = -18.63 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:02:35 2011