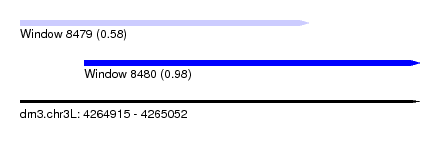
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 4,264,915 – 4,265,052 |
| Length | 137 |
| Max. P | 0.983093 |

| Location | 4,264,915 – 4,265,014 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 64.59 |
| Shannon entropy | 0.56974 |
| G+C content | 0.29729 |
| Mean single sequence MFE | -19.15 |
| Consensus MFE | -4.51 |
| Energy contribution | -6.70 |
| Covariance contribution | 2.19 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.24 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.584631 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
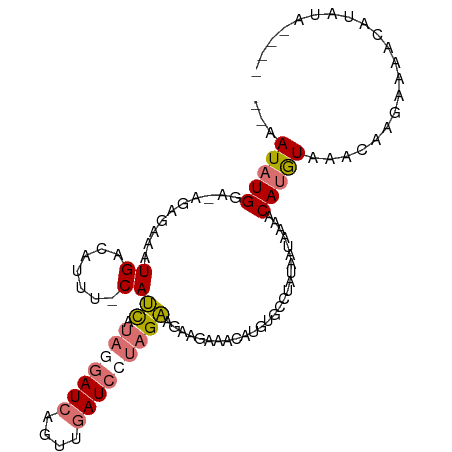
>dm3.chr3L 4264915 99 + 24543557 --AAUAUGGA-AGAGAAAUGUCAUUU-CAUCAUAAGAUCAGUUGAUCCUAGAUGAAGAAACAUGUGCCUAUAAUAAAACAUGUAAACAAGAAAACGUAUAUAA- --.(((((..-............(((-((((.((.((((....)))).)))))))))..((((((............))))))...........)))))....- ( -18.50, z-score = -2.34, R) >droSec1.super_2 4261989 98 + 7591821 CUAAUAUGGA-AGAGAAAUGACAUAU-CAUCAUAGGAUCAGUUGAUCCUAGAAGAAGAAACAUGUGCCUAUUAUAAAACAUGUAAACAAGAAAACAUAUA---- ...(((((..-..............(-(.((.(((((((....))))))))).))....((((((............))))))...........))))).---- ( -19.60, z-score = -2.82, R) >droSim1.chr3L 3805007 91 + 22553184 CUAAUAUGGA-AGAGAAAUGACAUUU-CAUCAUAGGAUCAGUUGAUCCUAGAAGAAGAAACAUGUGCCUAUUAUAAAACAUGUAAAACAUAUA----------- ...(((((..-............(((-(.((.(((((((....))))))))).))))..((((((............))))))....))))).----------- ( -21.10, z-score = -3.32, R) >droWil1.scaffold_180708 6332276 102 - 12563649 --AAUAUGUCCAGUCUAAUGACAGUUGCAAUCAACGAUUACAACAUAUCAGUAAUUAAAACGAUGGUAAAAACCAAUACAAAUGAAUCCACAGGUACGUAUAUA --.((((((((.(((....))).((((.((((...)))).))))...................((((....)))).................)).))))))... ( -17.40, z-score = -1.34, R) >consensus __AAUAUGGA_AGAGAAAUGACAUUU_CAUCAUAGGAUCAGUUGAUCCUAGAAGAAGAAACAUGUGCCUAUAAUAAAACAUGUAAACAAGAAAACAUAUA____ .............................((.(((((((....))))))))).......((((((............))))))..................... ( -4.51 = -6.70 + 2.19)

| Location | 4,264,937 – 4,265,052 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 74.77 |
| Shannon entropy | 0.32739 |
| G+C content | 0.28219 |
| Mean single sequence MFE | -17.97 |
| Consensus MFE | -13.87 |
| Energy contribution | -14.20 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.983093 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
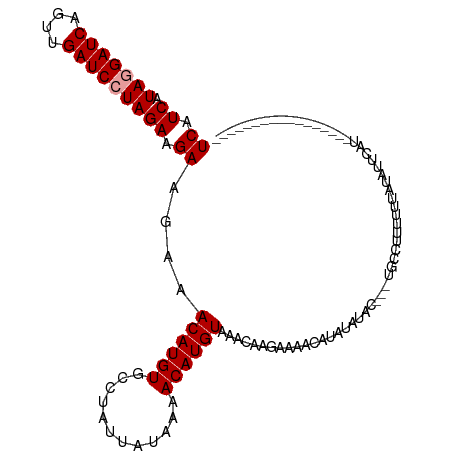
>dm3.chr3L 4264937 115 + 24543557 UCAUCAUAAGAUCAGUUGAUCCUAGAUGAAGAAACAUGUGCCUAUAAUAAAACAUGUAAACAAGAAAACGUAUAUAAACAUACAUACAUUAUAUUACCGCCUUUUUGUUAUUCAU (((((.((.((((....)))).))))))).(((((((((............)))))).((((((((..((.((((((...........))))))...))..)))))))).))).. ( -21.80, z-score = -3.27, R) >droSec1.super_2 4262013 95 + 7591821 UCAUCAUAGGAUCAGUUGAUCCUAGAAGAAGAAACAUGUGCCUAUUAUAAAACAUGUAAACAAGAAAACAUAUAUAC---UGCCAUUUUUAUAUUCAU----------------- ((.((.(((((((....))))))))).))....((((((............))))))....................---..................----------------- ( -16.00, z-score = -1.76, R) >droSim1.chr3L 3805031 88 + 22553184 UCAUCAUAGGAUCAGUUGAUCCUAGAAGAAGAAACAUGUGCCUAUUAUAAAACAUGUAAA-------ACAUAUAUAC---UGCCUUUUUUAUAUUCAU----------------- ......(((((((....)))))))(((((((..((((((............))))))...-------.((.......---)).)))))))........----------------- ( -16.10, z-score = -2.00, R) >consensus UCAUCAUAGGAUCAGUUGAUCCUAGAAGAAGAAACAUGUGCCUAUUAUAAAACAUGUAAACAAGAAAACAUAUAUAC___UGCCUUUUUUAUAUUCAU_________________ ((.((.(((((((....))))))))).))....((((((............)))))).......................................................... (-13.87 = -14.20 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:02:29 2011