
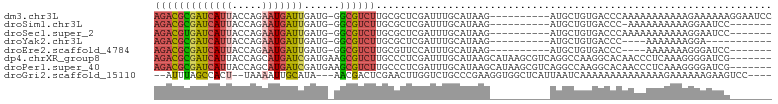
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 4,152,631 – 4,152,723 |
| Length | 92 |
| Max. P | 0.658318 |

| Location | 4,152,631 – 4,152,723 |
|---|---|
| Length | 92 |
| Sequences | 8 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 66.10 |
| Shannon entropy | 0.64428 |
| G+C content | 0.44723 |
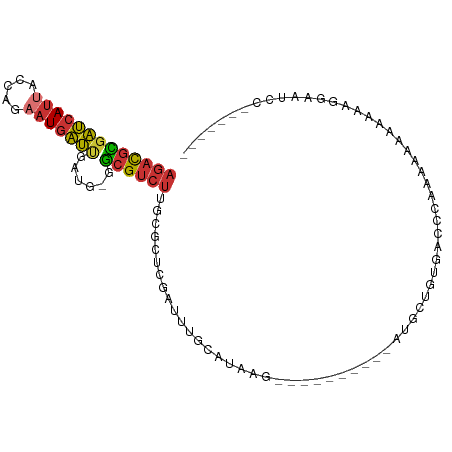
| Mean single sequence MFE | -21.69 |
| Consensus MFE | -8.42 |
| Energy contribution | -8.55 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.658318 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 4152631 92 - 24543557 AGACGCGAUCAUUACCAGAAUGAUUGAUG-GGCGUCUUGCGCUCGAUUUGCAUAAG----------AUGCUGUGACCCAAAAAAAAAAAGAAAAAAGGAAUCC ...((((((((((.....)))))))).))-(((((((((.((.......)).))))----------)))))................................ ( -20.90, z-score = -1.61, R) >droSim1.chr3L 3693419 84 - 22553184 AGACGCGAUCAUUACCAGAAUGAUUGAUG-GGCGUCUUGCGCUCGAUUUGCAUAAG----------AUGCUGUGACCC-AAAAAAAAAAGGAAUCC------- ...((((((((((.....)))))))).))-(((((((((.((.......)).))))----------))))).......-.................------- ( -20.90, z-score = -1.52, R) >droSec1.super_2 4145783 85 - 7591821 AGACGUGAUCAUUACCAGAAUGAUUGAUG-GGCGUCUUGCGCUCGAUUUGCAUAAG----------AUGCUGUGACCCAAAAAAAAAAAGGAAUCC------- ...((((((((((.....))))))).)))-(((((((((.((.......)).))))----------))))).........................------- ( -19.40, z-score = -1.21, R) >droYak2.chr3L 4723789 77 - 24197627 AGACGCGAUCAUUACCAGAAUGAUUGAUG-GGCGUCUUGCGCUCGAUUUGCAUAAG----------AUGCUGUGACCC----AAAAAAAGGA----------- ...((((((((((.....)))))))).))-(((((((((.((.......)).))))----------))))).....((----.......)).----------- ( -21.20, z-score = -1.65, R) >droEre2.scaffold_4784 6863104 81 - 25762168 AGACGCGAUCAUUACCAGAAUGAUUGAUG-GGCGUCUUGCGUUCCAUUUGCAUAAG----------AUGCUGUGACCC----AAAAAAAGGGAUCC------- ...((((((((((.....)))))))).))-(((((((((((.......))))..))----------)))))....(((----.......)))....------- ( -22.70, z-score = -1.72, R) >dp4.chrXR_group8 8315273 96 - 9212921 AGACGCGAUCAUUACCAGCAUGAUCGAUGAAGCGUCUUGCCCUCGAUUUGCAUAAGCAUAAGCGUCAGGCCAAGGCACAACCCUCAAAGGGGAUCG------- ...(((((((((.......))))))).))..(.((((((.(((((...(((....)))....))..))).)))))).)..((((....))))....------- ( -27.60, z-score = -1.21, R) >droPer1.super_40 506654 96 - 810552 AGACGCGAUCAUUACCAGCAUGAUCGAUGAAGCGUCUUGCCCUCGAUUUGCAUAAGCAUAAGCGUCAGGCCAAGGCACAACCCUCAAAGGGGAUCG------- ...(((((((((.......))))))).))..(.((((((.(((((...(((....)))....))..))).)))))).)..((((....))))....------- ( -27.60, z-score = -1.21, R) >droGri2.scaffold_15110 19503942 92 + 24565398 --AUUUAGCCACU--UAAAAUUGCAUA---AACGACUCGAACUUGGUCUGCCCGAAGGUGGCUCAUUAAUCAAAAAAAAAAAAAAGAAAAAAGAAGUCC---- --....(((((((--(......(((..---...((((.......)))))))....))))))))....................................---- ( -13.20, z-score = -0.38, R) >consensus AGACGCGAUCAUUACCAGAAUGAUUGAUG_GGCGUCUUGCGCUCGAUUUGCAUAAG__________AUGCUGUGACCCAAAAAAAAAAAGGAAUCC_______ (((((((((((((.....)))))))......)))))).................................................................. ( -8.42 = -8.55 + 0.13)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:02:15 2011