
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 4,144,615 – 4,144,717 |
| Length | 102 |
| Max. P | 0.585533 |

| Location | 4,144,615 – 4,144,717 |
|---|---|
| Length | 102 |
| Sequences | 9 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 59.71 |
| Shannon entropy | 0.76938 |
| G+C content | 0.56436 |
| Mean single sequence MFE | -29.87 |
| Consensus MFE | -9.93 |
| Energy contribution | -9.49 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.585533 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
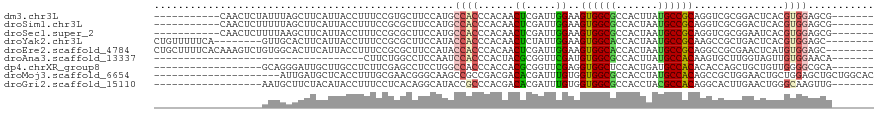
>dm3.chr3L 4144615 102 - 24543557 -----------CAACUCUAUUUAGCUUCAUUACCUUUCCGUGCUUCCAUGCCACCCACAACUCGAUUGGAAGUGGCGCCACUUAUGCCGCAGGUCGCGGACUCACGUGGAGCG------- -----------............(((((((......((((((...((..(((((...(((.....)))...)))))((..(....)..)).)).)))))).....))))))).------- ( -29.60, z-score = -1.51, R) >droSim1.chr3L 3684618 102 - 22553184 -----------CAACUCUUUUUAGCUUCAUUACCUUUCCGCGCUUCCAUGCCACCCACAACUCGAUUGGAAGUGGCGCCACUAAUGCCGCAGGUCGCGGACUCACGUGGAGCG------- -----------............(((((((......((((((...((..(((((...(((.....)))...)))))((..(....)..)).)).)))))).....))))))).------- ( -31.50, z-score = -2.04, R) >droSec1.super_2 4138057 102 - 7591821 -----------CAACUCUUUUAAGCUUCAUUACCUUUCCGCGCUUCCAUGCCACCCACAACUCGAUUGGAAGUGGCGCCACUAAUGCCGCAGGUCGCGGAAUCACGUGGAGCG------- -----------............(((((((.....(((((((...((..(((((...(((.....)))...)))))((..(....)..)).)).)))))))....))))))).------- ( -33.30, z-score = -2.57, R) >droYak2.chr3L 4715866 104 - 24197627 CUGUUUUUCA--------GUUGCACUUCAUUACCUUUCCGCGCUUCCAUACCACCCACAACUCUAUUGGAAGUGGCACCACUAAUGCCGCAAGCCGCUGACUCACGUGGAGC-------- .......(((--------((.((..................(((((((..................)))))))((((.......))))....)).)))))(((.....))).-------- ( -22.57, z-score = -0.57, R) >droEre2.scaffold_4784 6853477 112 - 25762168 CUGCUUUUCACAAAGUCUGUGGCACUUCAUUACCUUUCCGCGCUUCCAUACCACCCACAACUCGAUUGGAAGUGGCACCACUAAUGCCGCAGGCCGCGAACUCAUGUGGAGC-------- .....(((((((.(((.((((((.((.(.............(((((((..................)))))))((((.......))))).)))))))).)))..))))))).-------- ( -31.37, z-score = -1.40, R) >droAna3.scaffold_13337 14562192 78 - 23293914 -----------------------------------CUUCUGGCCUCCAAUCCACCCACUACGCGGUUCGAUGUGGCGCCACUUAUGCCACAAGUGCUUGGUAGUUGUGGAACA------- -----------------------------------....(((...))).(((((..(((((..(((.(..(((((((.......))))))).).)))..))))).)))))...------- ( -23.50, z-score = -0.67, R) >dp4.chrXR_group8 8308787 95 - 9212921 ------------------GCAGGGAUUGCUUGCCUCUUCGAGCCUCCUGGCCACCCACCACGCGGUUCGAGGUGGCUCCACUGAUGCCACACACCAGCUGCUGUUGGGGCGCA------- ------------------.((((((......(((.(((((((((.(.(((.......))).).))))))))).)))))).)))..(((.....(((((....))))))))...------- ( -37.90, z-score = -0.44, R) >droMoj3.scaffold_6654 727716 99 + 2564135 ---------------------AUUGAUGCUCACCUUUGCGAACGGGCAAGCCGCCGACGACACGAUUUGUGGUGGCGCCACCUAUGCCACAGCCGCUGGAACUGCUGGAGCUGCUGGCAC ---------------------.....(((........)))....(((..((((((.((((......))))))))))))).....(((((((((..(..(.....)..).)))).))))). ( -34.80, z-score = -0.62, R) >droGri2.scaffold_15110 3058710 95 - 24565398 ------------------AAUGCUUCUACAUACCUUUCCUCACAGGCAUACCGCCCACGACACGAUUUGUGGUGGCGCCACCUACGCCACAGGCACUUGAACUGGGCAAGUUG------- ------------------.((((((..................))))))...(((((...(((.....)))((((((.......))))))............)))))......------- ( -24.27, z-score = -0.47, R) >consensus __________________GUUGAGCUUCAUUACCUUUCCGCGCUUCCAUGCCACCCACAACUCGAUUGGAAGUGGCGCCACUAAUGCCGCAGGCCGCGGACUCACGUGGAGCG_______ ..................................................((((......((.....))..((((((.......))))))...............))))........... ( -9.93 = -9.49 + -0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:02:14 2011