| Sequence ID | dm3.chr3L |
|---|---|
| Location | 4,129,600 – 4,129,696 |
| Length | 96 |
| Max. P | 0.518667 |

| Location | 4,129,600 – 4,129,696 |
|---|---|
| Length | 96 |
| Sequences | 10 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 68.76 |
| Shannon entropy | 0.63552 |
| G+C content | 0.58708 |
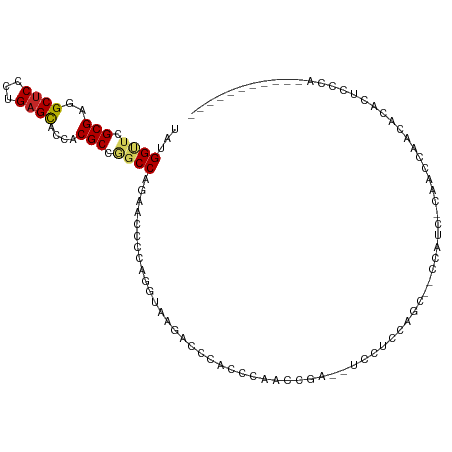
| Mean single sequence MFE | -18.03 |
| Consensus MFE | -10.86 |
| Energy contribution | -11.08 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.27 |
| Mean z-score | -0.23 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.518667 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
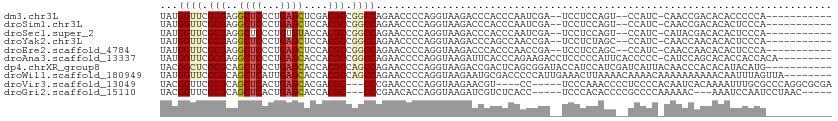
>dm3.chr3L 4129600 96 - 24543557 UAUGGUUCGCGAGGCUCCCUGAGCUCGACGCCGGCCAGAACCCCAGGUAAGACCCACCCAAUCGA--UCCUCCAGU--CCAUC-CAACCGACACACCCCCA----------- .((((...(.((((.((.(((.((.((....))))))).......(((.......))).....))--.)))))...--)))).-.................----------- ( -17.30, z-score = 0.93, R) >droSim1.chr3L 3669377 96 - 22553184 UAUGGUUCGCGAGGCUCCCUGAGCUCCACGCCGGCCAGAACCCCAGGUAAGACCCACCCAAUCGA--UCCUCCAGU--CCAUC-CAACCGACACACUCCCA----------- ..((((..(((.(((((...)))))...)))..))))........((...(((............--.......))--)...)-)................----------- ( -17.11, z-score = 0.74, R) >droSec1.super_2 4122062 96 - 7591821 UAUGGUUCGCGAGGCUCCCUGUGUACCACGCCGGCCAGAACCCCAGGUAAGACCCACCCAAUCGA--UCCUCCAGU--CCAUC-CAUACGACACACUCCCA----------- (((((...((((((.((((.(((.....))).))...........(((.......))).....))--.))))..))--....)-)))).............----------- ( -15.10, z-score = 1.61, R) >droYak2.chr3L 4700762 96 - 24197627 UAUGGUUCGCGAGGCUCCCUGAGCUCCACGCCGGCCAGAACCCCAGGUAAGACCCAGCCAACCGA--UCCUCUAGC--CCAUC-CAACCAACACACUCCCA----------- ..(((((.(((.(((((...)))))...))).(((.(((......(((..(......)..)))..--...))).))--)....-.)))))...........----------- ( -20.20, z-score = -0.52, R) >droEre2.scaffold_4784 6838581 96 - 25762168 UAUGGUUCGCGAGGCUCCCUGAGCUCCACGCCGGCCAGAACCCCAGGUAAGACCCACCCAACCGA--UCCUCCAGC--CCAUC-CAACCAACACACUCCCA----------- ..(((((.(((.(((((...)))))...))).(((..((......(((..(......)..)))..--...))..))--)....-.)))))...........----------- ( -18.30, z-score = -0.29, R) >droAna3.scaffold_13337 14550149 102 - 23293914 UAUGGUUCGCGAGGCUCCCUGAGCACCACGCCGGCCAGAACCCCAGGUAAGAUUCACCCAGAAGACCUCCCCCAUUCACCCCC-CAUCCAGCACACCACCACA--------- ..((((..(((..((((...))))....)))..))))(((....((((....(((.....))).))))......)))......-...................--------- ( -18.80, z-score = -0.52, R) >dp4.chrXR_group8 8295944 101 - 9212921 UACGGCUCGCGCAGCUCCCUGAGCACCACGCCAGCCAGAACCCCAGGUAAGACCGACUCAGCGGAUACCAUCCAUCGAUCAUUACAACCCACACAUACAUG----------- ...((((.(((..((((...))))....))).)))).........((((...(((......))).))))................................----------- ( -21.60, z-score = -1.92, R) >droWil1.scaffold_180949 4221615 104 - 6375548 UAUGGUUCGCGCAGCUCAUUGAGCACCACGCCAGCCAGAACCCCAGGUAAGAAUGCGACCCCCAUUGAAACUUAAAACAAAACAAAAAAAAAACAAUUUAGUUA-------- .((((....((((((((...)))).........(((.........))).....))))....)))).......................................-------- ( -14.50, z-score = -0.09, R) >droVir3.scaffold_13049 1983689 100 - 25233164 UACGGUUCGCGCAGCUCACUGAGCACGACGC---CCCGAACCCCAGGUAAGAACGU----CC-----UCCCAAACCCCUCCCCACAAUCACAAAAUUUGCGCCCAGGCGCGA ...(((((((((.((((...))))..).)))---...)))))...((..((...((----..-----......))..))..)).............((((((....)))))) ( -22.40, z-score = -0.80, R) >droGri2.scaffold_15110 3045555 96 - 24565398 UACGGUUCGCGCAGCUCACUGAGCACCACGC---CCCGAACACCAGGUAAGAUCGUCUCACC-----UCCCACACCCCGCCCCAAAAAC---AAAUCCAAUCCUAAC----- ..(((..((.(..((((...))))..).)).---.)))......((((.((.....)).)))-----).....................---...............----- ( -15.00, z-score = -1.46, R) >consensus UAUGGUUCGCGAGGCUCCCUGAGCACCACGCCGGCCAGAACCCCAGGUAAGACCCACCCAACCGA__UCCUCCAGC__CCAUC_CAACCAACACACUCCCA___________ ...((((.(((..((((...))))....))).))))............................................................................ (-10.86 = -11.08 + 0.22)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:02:13 2011