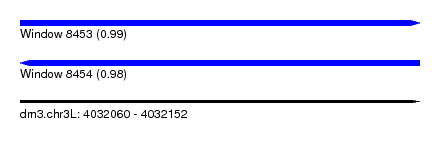
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 4,032,060 – 4,032,152 |
| Length | 92 |
| Max. P | 0.992093 |

| Location | 4,032,060 – 4,032,152 |
|---|---|
| Length | 92 |
| Sequences | 9 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 67.18 |
| Shannon entropy | 0.63894 |
| G+C content | 0.47771 |
| Mean single sequence MFE | -33.41 |
| Consensus MFE | -14.34 |
| Energy contribution | -13.84 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.77 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.52 |
| SVM RNA-class probability | 0.992093 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
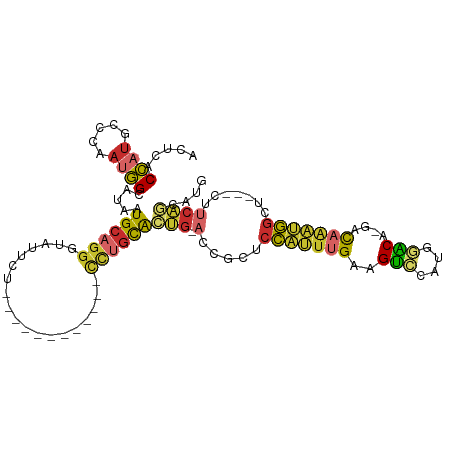
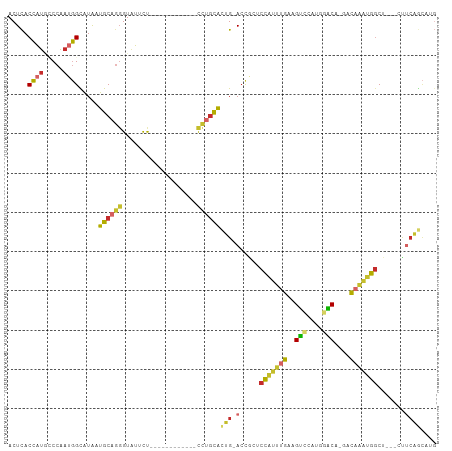
>dm3.chr3L 4032060 92 + 24543557 CAUGCUGAGG---AGCCAUUUGUC-UGUUCAUGGACUUCAAAUGGAGCGGU-CAGUGCAGG------------AGAAUACCCUGCAUUAUACCAUUGGGCAUGGUGAGU ((((((.(((---..(((((((..-.(((....)))..)))))))..)(((-.((((((((------------.......))))))))..))).)).))))))...... ( -35.60, z-score = -2.80, R) >droSim1.chr3L 3571822 90 + 22553184 CAUGCUGAG----AGCCAUUUGUC-UGUCCAUGGACUUCAAAUGGAGCGGU-CAGUGCAGG------------AGAAUACC-UGCAUUAUGCCAUUGGGCAUGGUGAGU ((((((.((----..(((((((..-.(((....)))..)))))))..)(((-.((((((((------------......))-))))))..)))..).))))))...... ( -37.80, z-score = -3.29, R) >droSec1.super_2 4024756 92 + 7591821 CAUGCUGAGG---AGCCAUUUGUC-UGUCCAUGGACUUCAAAUGGAGCGGU-CAGUGCAGG------------AGAAUACCCUGCAUUAUACCAUUGGGCAUGGUGAGU ((((((.(((---..(((((((..-.(((....)))..)))))))..)(((-.((((((((------------.......))))))))..))).)).))))))...... ( -37.90, z-score = -3.21, R) >droYak2.chr3L 4599703 92 + 24197627 CAUGUUGAAG---AGCCAUUUGUC-UGUCCAUGGACUUCAAAUGGAGCGGU-CAGUGCAGG------------AGAAUACCCUGCAUUAUACCCUUGGCAAUGGUGAGU (((((..(.(---..(((((((..-.(((....)))..)))))))..)(((-.((((((((------------.......))))))))..))).)..)).)))...... ( -32.10, z-score = -2.33, R) >droEre2.scaffold_4784 6736670 92 + 25762168 CAUGCUGAAG---AGCCAUUUGUC-UGUCCAGGGACUUCAAAUGGAGCGGU-CAGUGCAGG------------AGAAUACCCUGCAUUACACCAUUGGAAAUGGUGAGU ...(((((.(---..(((((((..-.(((....)))..)))))))..)..)-))))(((((------------.......)))))....((((((.....))))))... ( -36.30, z-score = -3.48, R) >droAna3.scaffold_13337 7073408 97 + 23293914 -------CAUUUUCGCCGGCAAUU-UGUUCAUGAACUUCAAAUGGAGCAGU-CCUUAUUAAAUCAUC---AGGAAAAUACCCUGCACCACGCCAUUGGCUAUGGUGAGU -------((((...((((((....-.(((....)))......(((.(((((-(((.((......)).---)))).......)))).))).)))...)))...))))... ( -20.31, z-score = 0.12, R) >dp4.chrXR_group6 6874638 107 - 13314419 CUUGCAGAAUGGUAGCCAUUUGUU-UGGCACCAGCCCUCAAAUGGAGUGGU-CCGCACAGAGUGUUCCAAAUAAGAACACCCUGCACCAUGCCAUUGGAUAUGGUGAGU ..(((....((((.((((......-))))))))((((((.....))).)))-..)))(((.((((((.......)))))).)))(((((((((...)).)))))))... ( -38.70, z-score = -2.67, R) >droPer1.super_82 158337 107 + 267963 CUUGCAGAAUGUUAGCCAUUUGUU-UGGCACCAGCCCUCAAAUGGAGUGGU-CCGCACAGAGUGUUCCAAAUAGGAAUACCCUGCACCAUGCCAUUGGAUAUGGUGAGU ..(((.((.......(((((((..-.(((....)))..))))))).....)-).)))(((.(((((((.....))))))).)))(((((((((...)).)))))))... ( -38.20, z-score = -2.73, R) >droWil1.scaffold_180698 9643590 98 + 11422946 --CAUUGCGGGUUGGGUGAUAAUUGUGAACAUUACCUUCAUUCAAAUUGUUGCAAUGCCAG---------AGCGAAAUACACUGCAACAUGCAUUGGGUCCCUGUAAGU --..(((((((((((((((....((....))......)))))))).......((((((...---------.(((........))).....))))))....))))))).. ( -23.80, z-score = 0.31, R) >consensus CAUGCUGAAG___AGCCAUUUGUC_UGUCCAUGGACUUCAAAUGGAGCGGU_CAGUGCAGG____________AGAAUACCCUGCAUUAUGCCAUUGGACAUGGUGAGU ..(((..........(((((((....(((....)))..))))))).(((......))).........................)))...((((((.....))))))... (-14.34 = -13.84 + -0.50)

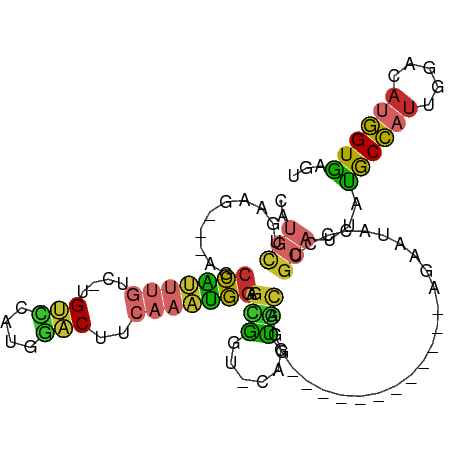
| Location | 4,032,060 – 4,032,152 |
|---|---|
| Length | 92 |
| Sequences | 9 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 67.18 |
| Shannon entropy | 0.63894 |
| G+C content | 0.47771 |
| Mean single sequence MFE | -29.57 |
| Consensus MFE | -12.19 |
| Energy contribution | -11.63 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.65 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.977668 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 4032060 92 - 24543557 ACUCACCAUGCCCAAUGGUAUAAUGCAGGGUAUUCU------------CCUGCACUG-ACCGCUCCAUUUGAAGUCCAUGAACA-GACAAAUGGCU---CCUCAGCAUG ......(((((.....(((....(((((((.....)------------))))))...-)))...(((((((..((......)).-..)))))))..---.....))))) ( -24.40, z-score = -1.42, R) >droSim1.chr3L 3571822 90 - 22553184 ACUCACCAUGCCCAAUGGCAUAAUGCA-GGUAUUCU------------CCUGCACUG-ACCGCUCCAUUUGAAGUCCAUGGACA-GACAAAUGGCU----CUCAGCAUG .......(((((....)))))..((((-((......------------))))))(((-(.....(((((((..(((....))).-..)))))))..----.)))).... ( -28.50, z-score = -2.53, R) >droSec1.super_2 4024756 92 - 7591821 ACUCACCAUGCCCAAUGGUAUAAUGCAGGGUAUUCU------------CCUGCACUG-ACCGCUCCAUUUGAAGUCCAUGGACA-GACAAAUGGCU---CCUCAGCAUG ......(((((.....(((....(((((((.....)------------))))))...-)))...(((((((..(((....))).-..)))))))..---.....))))) ( -29.30, z-score = -2.38, R) >droYak2.chr3L 4599703 92 - 24197627 ACUCACCAUUGCCAAGGGUAUAAUGCAGGGUAUUCU------------CCUGCACUG-ACCGCUCCAUUUGAAGUCCAUGGACA-GACAAAUGGCU---CUUCAACAUG ................(((....(((((((.....)------------))))))...-)))...(((((((..(((....))).-..)))))))..---.......... ( -23.90, z-score = -0.94, R) >droEre2.scaffold_4784 6736670 92 - 25762168 ACUCACCAUUUCCAAUGGUGUAAUGCAGGGUAUUCU------------CCUGCACUG-ACCGCUCCAUUUGAAGUCCCUGGACA-GACAAAUGGCU---CUUCAGCAUG ...((((((.....))))))...(((((((.....)------------))))))(((-(.....(((((((..(((....))).-..)))))))..---..)))).... ( -29.60, z-score = -2.70, R) >droAna3.scaffold_13337 7073408 97 - 23293914 ACUCACCAUAGCCAAUGGCGUGGUGCAGGGUAUUUUCCU---GAUGAUUUAAUAAGG-ACUGCUCCAUUUGAAGUUCAUGAACA-AAUUGCCGGCGAAAAUG------- .((((((((.(((...))))))))).))..(((((((((---(..(((((.....((-(....))).......(((....))))-))))..))).)))))))------- ( -24.70, z-score = -0.79, R) >dp4.chrXR_group6 6874638 107 + 13314419 ACUCACCAUAUCCAAUGGCAUGGUGCAGGGUGUUCUUAUUUGGAACACUCUGUGCGG-ACCACUCCAUUUGAGGGCUGGUGCCA-AACAAAUGGCUACCAUUCUGCAAG ...((((((..(....)..))))))(((((((((((.....)))))))))))(((((-(((.(((.....))))).((((((((-......)))).)))).)))))).. ( -43.50, z-score = -3.96, R) >droPer1.super_82 158337 107 - 267963 ACUCACCAUAUCCAAUGGCAUGGUGCAGGGUAUUCCUAUUUGGAACACUCUGUGCGG-ACCACUCCAUUUGAGGGCUGGUGCCA-AACAAAUGGCUAACAUUCUGCAAG ...((((((..(....)..))))))((((((.((((.....)))).))))))(((((-(.....(((((((..(((....))).-..))))))).......)))))).. ( -39.00, z-score = -3.09, R) >droWil1.scaffold_180698 9643590 98 - 11422946 ACUUACAGGGACCCAAUGCAUGUUGCAGUGUAUUUCGCU---------CUGGCAUUGCAACAAUUUGAAUGAAGGUAAUGUUCACAAUUAUCACCCAACCCGCAAUG-- .......(((...(((....(((((((((((........---------...)))))))))))..)))......((((((.......)))))).)))...........-- ( -23.20, z-score = -0.59, R) >consensus ACUCACCAUGCCCAAUGGCAUAAUGCAGGGUAUUCU____________CCUGCACUG_ACCGCUCCAUUUGAAGUCCAUGGACA_GACAAAUGGCU___CUUCAGCAUG .....((((.....)))).....((((((...................))))))..........(((((((..(((....)))....)))))))............... (-12.19 = -11.63 + -0.56)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:02:06 2011