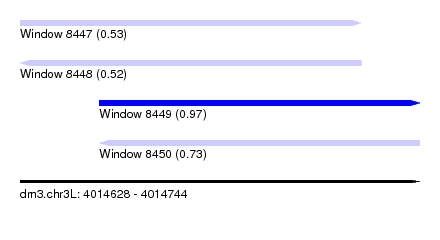
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 4,014,628 – 4,014,744 |
| Length | 116 |
| Max. P | 0.969270 |

| Location | 4,014,628 – 4,014,727 |
|---|---|
| Length | 99 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 82.55 |
| Shannon entropy | 0.35441 |
| G+C content | 0.45452 |
| Mean single sequence MFE | -29.72 |
| Consensus MFE | -19.72 |
| Energy contribution | -19.81 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.531699 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
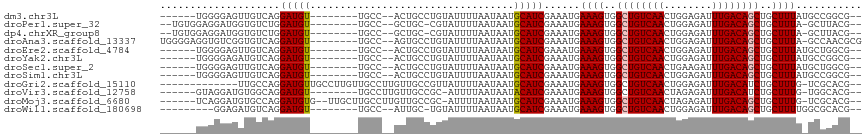
>dm3.chr3L 4014628 99 + 24543557 ------UGGGGAGUUGUCAGGAUGU--------UGCC--ACUGCCUGUAUUUUAAUAAUGCAUCGAAAUGAAAGUGGCUGUCAACUGGAGAUUUGACAGCUGCUUUAUGCCGGCG-- ------.(((.(((.(.(((....)--------))).--))).)))(((((.....))))).(((..(((((.(..((((((((........))))))))..))))))..)))..-- ( -29.60, z-score = -1.23, R) >droPer1.super_32 169444 101 - 992631 --UGUGGAGGAUGGUGUCUGGAUGU--------UGCC--GCUGC-CGUAUUUUAAUAAUGCAUCGAAAUGAAAGUGGCUGUCAACUGGAGAUUUGACAGCUGCUUUA-GCUUACG-- --...((.((.(((((.(.....).--------))))--))).)-)(((((.....))))).........((((..((((((((........))))))))..)))).-.......-- ( -28.30, z-score = -1.14, R) >dp4.chrXR_group8 661462 101 + 9212921 --UGUGGAGGAUGGUGUCUGGAUGU--------UGCC--GCUGC-CGUAUUUUAAUAAUGCAUCGAAAUGAAAGUGGCUGUCAACUGGAGAUUUGACAGCUGCUUUA-GCUUACG-- --...((.((.(((((.(.....).--------))))--))).)-)(((((.....))))).........((((..((((((((........))))))))..)))).-.......-- ( -28.30, z-score = -1.14, R) >droAna3.scaffold_13337 1867696 106 + 23293914 UGGGGAGGUGUCGGUGUCAGGAUGU--------UGCC--AGUGCCUGUAUUUUAAUAAUGCAUCGAAAUGAAAGUGGCUGUCAACUGGAGAUUUGACAGCUGCUUUA-GCCAACGCG .(.(..(((.(((((((((((.((.--------...)--)...))))............)))))))....((((..((((((((........))))))))..)))).-)))..).). ( -35.10, z-score = -1.87, R) >droEre2.scaffold_4784 6718937 99 + 25762168 ------UGGGGAGUUGUCAGGAUGU--------UGCC--ACUGCCUGUAUUUUAAUAAUGCAUCGAAAUGAAAGUGGCUGUCAACUGGAGAUUUGACAGCUGCUUUAUGCUGGCG-- ------.........(((((.((..--------....--......((((((.....))))))........((((..((((((((........))))))))..)))))).))))).-- ( -29.50, z-score = -1.41, R) >droYak2.chr3L 4581632 99 + 24197627 ------UGGGGAGAUGUCAGGAUGU--------UGCC--ACUGCCUGUAUUUUAAUAAUGCAUCGAAAUGAAAGUGGCUGUCAACUGGAGAUUUGACAGCUGCUUUAUGCCGGCG-- ------(((.((.((......)).)--------).))--)..(((((((((.....))))))........((((..((((((((........))))))))..)))).....))).-- ( -31.50, z-score = -1.99, R) >droSec1.super_2 4007611 99 + 7591821 ------UGGGGAGUUGUCAGGAUGU--------UGCC--ACUGCCUGUAUUUUAAUAAUGCAUCGAAAUGAAAGUGGCUGUCAACUGAAGAUUUGACAGCUGCUUUAUGCUGGCG-- ------.........(((((.((..--------....--......((((((.....))))))........((((..((((((((........))))))))..)))))).))))).-- ( -29.50, z-score = -1.67, R) >droSim1.chr3L 3553520 99 + 22553184 ------UGGGGAGUUGUCAGGAUGU--------UGCC--ACUGCCUGUAUUUUAAUAAUGCAUCGAAAUGAAAGUGGCUGUCAACUGGAGAUUUGACAGCUGCUUUAUGCCGGCG-- ------.(((.(((.(.(((....)--------))).--))).)))(((((.....))))).(((..(((((.(..((((((((........))))))))..))))))..)))..-- ( -29.60, z-score = -1.23, R) >droGri2.scaffold_15110 563325 101 + 24565398 -------------UUGCCAGGAUGUUGCCUUGUUGCCUUGUUGCCGUUAUUUUAAUAAUGCAUCGAAAUGAAAGUGGCUGUCAACUGGAGAUUUGACAUCUGCUUUG-UCGCACG-- -------------......(((((((.((..((((.(..((..(..((((((..((.....))..))))))..)..)).).)))).))......)))))))((....-..))...-- ( -21.40, z-score = 0.62, R) >droVir3.scaffold_12758 21283 99 - 972128 ------GUAGGAUGUGGCAGGAUGU--------UGCCUUGUUGCCGC-AUUUUAAUAAUACAUCGAAAUGAAAGUGGCUGUCAACUAGAGAUUUGACAUCUGCUUUG-UGGCACG-- ------.(((((((((((((.(...--------.....).)))))))-))))))..........(..((.((((..(.((((((........)))))).)..)))))-)..)...-- ( -29.00, z-score = -1.64, R) >droMoj3.scaffold_6680 22762808 105 - 24764193 ------UCAGGAUGUGCCAGGAUGUG--UUGCUUGCCUUGUUGCCGC-AUUUUAAUAAUGCAUCGAAAUGAAAGUGGCUGUCAACUAGAGAUUUGACAGCUGCUUUG-UCGCACG-- ------.......((((...((((((--(((.(((...(((....))-)...))))))))))))((....((((..((((((((........))))))))..)))).-)))))).-- ( -33.50, z-score = -1.72, R) >droWil1.scaffold_180698 8787204 95 + 11422946 ---------GGAGAUGUCAGGAUGU--------UGCC--AUUGC-UGUAUUUUAAUAAUGCAUCGAAAUGAAAGUGGCUGUCAACUGGAGAUUUGACAGCUGCUUUUGGCGCACG-- ---------..............((--------((((--.(((.-((((((.....)))))).)))...(((((..((((((((........))))))))..))))))))).)).-- ( -31.40, z-score = -2.61, R) >consensus ______UGGGGAGGUGUCAGGAUGU________UGCC__ACUGCCUGUAUUUUAAUAAUGCAUCGAAAUGAAAGUGGCUGUCAACUGGAGAUUUGACAGCUGCUUUA_GCCGACG__ ....................(((((..................................)))))......((((..((((((((........))))))))..))))........... (-19.72 = -19.81 + 0.09)
| Location | 4,014,628 – 4,014,727 |
|---|---|
| Length | 99 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 82.55 |
| Shannon entropy | 0.35441 |
| G+C content | 0.45452 |
| Mean single sequence MFE | -22.77 |
| Consensus MFE | -13.23 |
| Energy contribution | -13.24 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.515555 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 4014628 99 - 24543557 --CGCCGGCAUAAAGCAGCUGUCAAAUCUCCAGUUGACAGCCACUUUCAUUUCGAUGCAUUAUUAAAAUACAGGCAGU--GGCA--------ACAUCCUGACAACUCCCCA------ --.(((((.((((((..((((((((........))))))))..)))).)).)))..))............((((..((--(...--------.)))))))...........------ ( -24.80, z-score = -2.22, R) >droPer1.super_32 169444 101 + 992631 --CGUAAGC-UAAAGCAGCUGUCAAAUCUCCAGUUGACAGCCACUUUCAUUUCGAUGCAUUAUUAAAAUACG-GCAGC--GGCA--------ACAUCCAGACACCAUCCUCCACA-- --.....((-.((((..((((((((........))))))))..)))).....((.(((..(((....)))..-))).)--))).--------.......................-- ( -18.10, z-score = -0.94, R) >dp4.chrXR_group8 661462 101 - 9212921 --CGUAAGC-UAAAGCAGCUGUCAAAUCUCCAGUUGACAGCCACUUUCAUUUCGAUGCAUUAUUAAAAUACG-GCAGC--GGCA--------ACAUCCAGACACCAUCCUCCACA-- --.....((-.((((..((((((((........))))))))..)))).....((.(((..(((....)))..-))).)--))).--------.......................-- ( -18.10, z-score = -0.94, R) >droAna3.scaffold_13337 1867696 106 - 23293914 CGCGUUGGC-UAAAGCAGCUGUCAAAUCUCCAGUUGACAGCCACUUUCAUUUCGAUGCAUUAUUAAAAUACAGGCACU--GGCA--------ACAUCCUGACACCGACACCUCCCCA .(((((((.-.((((..((((((((........))))))))..))))....)))))))............((((...(--(...--------.)).))))................. ( -27.90, z-score = -2.80, R) >droEre2.scaffold_4784 6718937 99 - 25762168 --CGCCAGCAUAAAGCAGCUGUCAAAUCUCCAGUUGACAGCCACUUUCAUUUCGAUGCAUUAUUAAAAUACAGGCAGU--GGCA--------ACAUCCUGACAACUCCCCA------ --.....((((((((..((((((((........))))))))..)))).......))))............((((..((--(...--------.)))))))...........------ ( -24.31, z-score = -2.62, R) >droYak2.chr3L 4581632 99 - 24197627 --CGCCGGCAUAAAGCAGCUGUCAAAUCUCCAGUUGACAGCCACUUUCAUUUCGAUGCAUUAUUAAAAUACAGGCAGU--GGCA--------ACAUCCUGACAUCUCCCCA------ --.(((((.((((((..((((((((........))))))))..)))).)).)))..))............((((..((--(...--------.)))))))...........------ ( -24.80, z-score = -2.21, R) >droSec1.super_2 4007611 99 - 7591821 --CGCCAGCAUAAAGCAGCUGUCAAAUCUUCAGUUGACAGCCACUUUCAUUUCGAUGCAUUAUUAAAAUACAGGCAGU--GGCA--------ACAUCCUGACAACUCCCCA------ --.....((((((((..((((((((........))))))))..)))).......))))............((((..((--(...--------.)))))))...........------ ( -24.31, z-score = -2.58, R) >droSim1.chr3L 3553520 99 - 22553184 --CGCCGGCAUAAAGCAGCUGUCAAAUCUCCAGUUGACAGCCACUUUCAUUUCGAUGCAUUAUUAAAAUACAGGCAGU--GGCA--------ACAUCCUGACAACUCCCCA------ --.(((((.((((((..((((((((........))))))))..)))).)).)))..))............((((..((--(...--------.)))))))...........------ ( -24.80, z-score = -2.22, R) >droGri2.scaffold_15110 563325 101 - 24565398 --CGUGCGA-CAAAGCAGAUGUCAAAUCUCCAGUUGACAGCCACUUUCAUUUCGAUGCAUUAUUAAAAUAACGGCAACAAGGCAACAAGGCAACAUCCUGGCAA------------- --..(((..-....)))..((((((........))))))((((...((.....))(((..............(....)..(....)...)))......))))..------------- ( -18.30, z-score = 0.33, R) >droVir3.scaffold_12758 21283 99 + 972128 --CGUGCCA-CAAAGCAGAUGUCAAAUCUCUAGUUGACAGCCACUUUCAUUUCGAUGUAUUAUUAAAAU-GCGGCAACAAGGCA--------ACAUCCUGCCACAUCCUAC------ --..((((.-.((((....((((((........))))))....)))).........(((((.....)))-))))))....((((--------......)))).........------ ( -20.90, z-score = -1.47, R) >droMoj3.scaffold_6680 22762808 105 + 24764193 --CGUGCGA-CAAAGCAGCUGUCAAAUCUCUAGUUGACAGCCACUUUCAUUUCGAUGCAUUAUUAAAAU-GCGGCAACAAGGCAAGCAA--CACAUCCUGGCACAUCCUGA------ --.((((((-((.......)))).........((((...(((..............(((((.....)))-))(....)..)))...)))--)........)))).......------ ( -25.70, z-score = -0.89, R) >droWil1.scaffold_180698 8787204 95 - 11422946 --CGUGCGCCAAAAGCAGCUGUCAAAUCUCCAGUUGACAGCCACUUUCAUUUCGAUGCAUUAUUAAAAUACA-GCAAU--GGCA--------ACAUCCUGACAUCUCC--------- --.((..((((((((..((((((((........))))))))..))))........(((..(((....)))..-))).)--))).--------))..............--------- ( -21.20, z-score = -1.88, R) >consensus __CGUCAGC_UAAAGCAGCUGUCAAAUCUCCAGUUGACAGCCACUUUCAUUUCGAUGCAUUAUUAAAAUACAGGCAGU__GGCA________ACAUCCUGACAACUCCCCA______ ...((......((((..((((((((........))))))))..))))........(((..(((....)))...))).....)).................................. (-13.23 = -13.24 + 0.02)
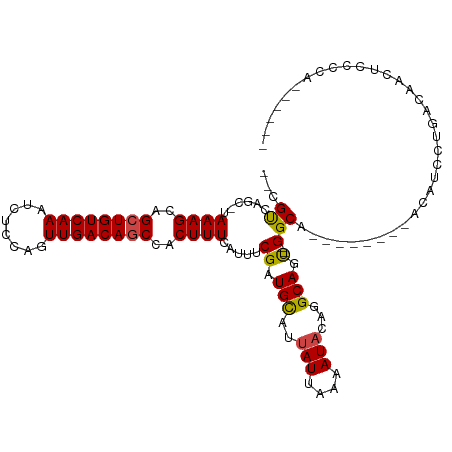
| Location | 4,014,651 – 4,014,744 |
|---|---|
| Length | 93 |
| Sequences | 11 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 90.56 |
| Shannon entropy | 0.18798 |
| G+C content | 0.42990 |
| Mean single sequence MFE | -26.82 |
| Consensus MFE | -23.25 |
| Energy contribution | -23.44 |
| Covariance contribution | 0.18 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.969270 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 4014651 93 + 24543557 -ACUGCCUGUAUUUUAAUAAUGCAUCGAAAUGAAAGUGGCUGUCAACUGGAGAUUUGACAGCUGCUUUAUGCCG-GCGACAAGGAUCGAUGAUGG---- -.....................((((((....((((..((((((((........))))))))..))))...((.-.......)).))))))....---- ( -27.60, z-score = -2.47, R) >droPer1.super_32 169471 91 - 992631 -GCUGCC-GUAUUUUAAUAAUGCAUCGAAAUGAAAGUGGCUGUCAACUGGAGAUUUGACAGCUGCUUUAGCUUA--CGACAAGGAUCGAUGAUGG---- -....((-(((((.....)))))(((((....((((..((((((((........))))))))..))))..(((.--....)))..)))))...))---- ( -26.00, z-score = -2.34, R) >dp4.chrXR_group8 661489 91 + 9212921 -GCUGCC-GUAUUUUAAUAAUGCAUCGAAAUGAAAGUGGCUGUCAACUGGAGAUUUGACAGCUGCUUUAGCUUA--CGACAAGGAUCGAUGAUGG---- -....((-(((((.....)))))(((((....((((..((((((((........))))))))..))))..(((.--....)))..)))))...))---- ( -26.00, z-score = -2.34, R) >droAna3.scaffold_13337 1867725 98 + 23293914 -AGUGCCUGUAUUUUAAUAAUGCAUCGAAAUGAAAGUGGCUGUCAACUGGAGAUUUGACAGCUGCUUUAGCCAACGCGACAAGGAUCGAUGAUGGAAGG -....((((((((.....)))))(((((....((((..((((((((........))))))))..)))).((....))........)))))......))) ( -28.00, z-score = -2.00, R) >droEre2.scaffold_4784 6718960 93 + 25762168 -ACUGCCUGUAUUUUAAUAAUGCAUCGAAAUGAAAGUGGCUGUCAACUGGAGAUUUGACAGCUGCUUUAUGCUG-GCGACAAGGAUCGAUGAUGG---- -.....................((((((....((((..((((((((........))))))))..))))....((-....))....))))))....---- ( -27.20, z-score = -2.56, R) >droYak2.chr3L 4581655 93 + 24197627 -ACUGCCUGUAUUUUAAUAAUGCAUCGAAAUGAAAGUGGCUGUCAACUGGAGAUUUGACAGCUGCUUUAUGCCG-GCGACAAGGAUCGAUGAUGG---- -.....................((((((....((((..((((((((........))))))))..))))...((.-.......)).))))))....---- ( -27.60, z-score = -2.47, R) >droSec1.super_2 4007634 93 + 7591821 -ACUGCCUGUAUUUUAAUAAUGCAUCGAAAUGAAAGUGGCUGUCAACUGAAGAUUUGACAGCUGCUUUAUGCUG-GCGACAAGGAUCGAUGAUGG---- -.....................((((((....((((..((((((((........))))))))..))))....((-....))....))))))....---- ( -27.20, z-score = -2.84, R) >droSim1.chr3L 3553543 93 + 22553184 -ACUGCCUGUAUUUUAAUAAUGCAUCGAAAUGAAAGUGGCUGUCAACUGGAGAUUUGACAGCUGCUUUAUGCCG-GCGACAAGGAUCGAUGAUGG---- -.....................((((((....((((..((((((((........))))))))..))))...((.-.......)).))))))....---- ( -27.60, z-score = -2.47, R) >droGri2.scaffold_15110 563350 93 + 24565398 UGUUGCCGUUAUUUUAAUAAUGCAUCGAAAUGAAAGUGGCUGUCAACUGGAGAUUUGACAUCUGCUUUGUCGCA--CGACAAGGAUCGAUGAUGG---- ......((((((....))))))((((((....((((..(.((((((........)))))).)..))))(((...--.))).....))))))....---- ( -24.00, z-score = -1.11, R) >droVir3.scaffold_12758 21307 92 - 972128 UGUUGCCGC-AUUUUAAUAAUACAUCGAAAUGAAAGUGGCUGUCAACUAGAGAUUUGACAUCUGCUUUGUGGCA--CGACAAGGAUCGAUGAUGG---- ...((((((-((((..((.....))..)))).((((..(.((((((........)))))).)..))))))))))--(((......))).......---- ( -22.90, z-score = -1.23, R) >droMoj3.scaffold_6680 22762838 91 - 24764193 UGUUGCCGC-AUUUUAAUAAUGCAUCGAAAUGAAAGUGGCUGUCAACUAGAGAUUUGACAGCUGCUUUGUCGCA--CGACAAGGAUCGAUGAUG----- .......((-(((.....)))))(((((....((((..((((((((........))))))))..))))(((...--.))).....)))))....----- ( -30.90, z-score = -3.38, R) >consensus _ACUGCCUGUAUUUUAAUAAUGCAUCGAAAUGAAAGUGGCUGUCAACUGGAGAUUUGACAGCUGCUUUAUGCCA_GCGACAAGGAUCGAUGAUGG____ ......................((((((....((((..((((((((........))))))))..))))........(.....)..))))))........ (-23.25 = -23.44 + 0.18)

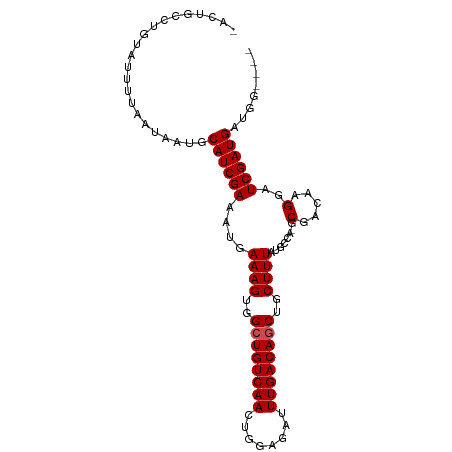
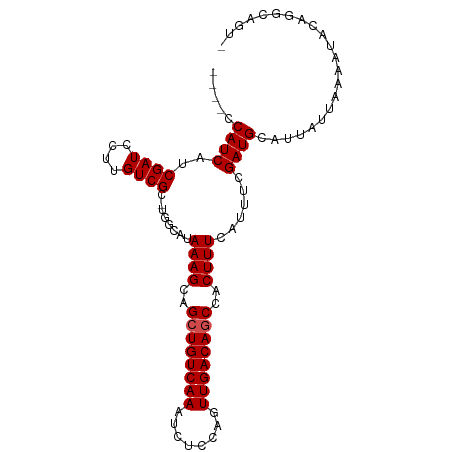
| Location | 4,014,651 – 4,014,744 |
|---|---|
| Length | 93 |
| Sequences | 11 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 90.56 |
| Shannon entropy | 0.18798 |
| G+C content | 0.42990 |
| Mean single sequence MFE | -20.22 |
| Consensus MFE | -16.14 |
| Energy contribution | -16.32 |
| Covariance contribution | 0.18 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.732981 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 4014651 93 - 24543557 ----CCAUCAUCGAUCCUUGUCGC-CGGCAUAAAGCAGCUGUCAAAUCUCCAGUUGACAGCCACUUUCAUUUCGAUGCAUUAUUAAAAUACAGGCAGU- ----....((((((....(((...-..))).((((..((((((((........))))))))..))))....)))))).....................- ( -20.20, z-score = -1.70, R) >droPer1.super_32 169471 91 + 992631 ----CCAUCAUCGAUCCUUGUCG--UAAGCUAAAGCAGCUGUCAAAUCUCCAGUUGACAGCCACUUUCAUUUCGAUGCAUUAUUAAAAUAC-GGCAGC- ----.............((((((--((.((....)).((((((((........))))))))...........................)))-))))).- ( -21.00, z-score = -2.54, R) >dp4.chrXR_group8 661489 91 - 9212921 ----CCAUCAUCGAUCCUUGUCG--UAAGCUAAAGCAGCUGUCAAAUCUCCAGUUGACAGCCACUUUCAUUUCGAUGCAUUAUUAAAAUAC-GGCAGC- ----.............((((((--((.((....)).((((((((........))))))))...........................)))-))))).- ( -21.00, z-score = -2.54, R) >droAna3.scaffold_13337 1867725 98 - 23293914 CCUUCCAUCAUCGAUCCUUGUCGCGUUGGCUAAAGCAGCUGUCAAAUCUCCAGUUGACAGCCACUUUCAUUUCGAUGCAUUAUUAAAAUACAGGCACU- ....((......(((....)))(((((((..((((..((((((((........))))))))..))))....)))))))..............))....- ( -22.10, z-score = -2.01, R) >droEre2.scaffold_4784 6718960 93 - 25762168 ----CCAUCAUCGAUCCUUGUCGC-CAGCAUAAAGCAGCUGUCAAAUCUCCAGUUGACAGCCACUUUCAUUUCGAUGCAUUAUUAAAAUACAGGCAGU- ----........(((....)))((-(.((((((((..((((((((........))))))))..)))).......))))(((.....)))...)))...- ( -19.31, z-score = -1.97, R) >droYak2.chr3L 4581655 93 - 24197627 ----CCAUCAUCGAUCCUUGUCGC-CGGCAUAAAGCAGCUGUCAAAUCUCCAGUUGACAGCCACUUUCAUUUCGAUGCAUUAUUAAAAUACAGGCAGU- ----....((((((....(((...-..))).((((..((((((((........))))))))..))))....)))))).....................- ( -20.20, z-score = -1.70, R) >droSec1.super_2 4007634 93 - 7591821 ----CCAUCAUCGAUCCUUGUCGC-CAGCAUAAAGCAGCUGUCAAAUCUUCAGUUGACAGCCACUUUCAUUUCGAUGCAUUAUUAAAAUACAGGCAGU- ----........(((....)))((-(.((((((((..((((((((........))))))))..)))).......))))(((.....)))...)))...- ( -19.31, z-score = -1.89, R) >droSim1.chr3L 3553543 93 - 22553184 ----CCAUCAUCGAUCCUUGUCGC-CGGCAUAAAGCAGCUGUCAAAUCUCCAGUUGACAGCCACUUUCAUUUCGAUGCAUUAUUAAAAUACAGGCAGU- ----....((((((....(((...-..))).((((..((((((((........))))))))..))))....)))))).....................- ( -20.20, z-score = -1.70, R) >droGri2.scaffold_15110 563350 93 - 24565398 ----CCAUCAUCGAUCCUUGUCG--UGCGACAAAGCAGAUGUCAAAUCUCCAGUUGACAGCCACUUUCAUUUCGAUGCAUUAUUAAAAUAACGGCAACA ----....((((((...(((((.--...))))).((...((((((........))))))))..........))))))...............(....). ( -17.40, z-score = -0.81, R) >droVir3.scaffold_12758 21307 92 + 972128 ----CCAUCAUCGAUCCUUGUCG--UGCCACAAAGCAGAUGUCAAAUCUCUAGUUGACAGCCACUUUCAUUUCGAUGUAUUAUUAAAAU-GCGGCAACA ----.......((((....))))--((((..((((....((((((........))))))....)))).........(((((.....)))-))))))... ( -16.40, z-score = -0.45, R) >droMoj3.scaffold_6680 22762838 91 + 24764193 -----CAUCAUCGAUCCUUGUCG--UGCGACAAAGCAGCUGUCAAAUCUCUAGUUGACAGCCACUUUCAUUUCGAUGCAUUAUUAAAAU-GCGGCAACA -----....(((((.....(((.--...)))((((..((((((((........))))))))..))))....)))))(((((.....)))-))(....). ( -25.30, z-score = -2.81, R) >consensus ____CCAUCAUCGAUCCUUGUCGC_UGGCAUAAAGCAGCUGUCAAAUCUCCAGUUGACAGCCACUUUCAUUUCGAUGCAUUAUUAAAAUACAGGCAGU_ ........((((((....((...........((((..((((((((........))))))))..))))))..))))))...................... (-16.14 = -16.32 + 0.18)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:02:03 2011